Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas
- PMID: 23826111
- PMCID: PMC3691184
- DOI: 10.1371/journal.pone.0066676
Integrated miRNA-mRNA analysis revealing the potential roles of miRNAs in chordomas
Abstract
Introduction: Emerging evidence suggests that microRNAs (miRNAs) are crucially involved in tumorigenesis and that paired expression profiles of miRNAs and mRNAs can be used to identify functional miRNA-target relationships with high precision. However, no studies have applied integrated analysis to miRNA and mRNA profiles in chordomas. The purpose of this study was to provide insights into the pathogenesis of chordomas by using this integrated analysis method.
Methods: Differentially expressed miRNAs and mRNAs of chordomas (n = 3) and notochord tissues (n = 3) were analyzed by using microarrays with hierarchical clustering analysis. Subsequently, the target genes of the differentially expressed miRNAs were predicted and overlapped with the differentially expressed mRNAs. Then, GO and pathway analyses were performed for the intersecting genes.
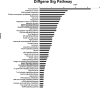
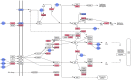
Results: The microarray analysis indicated that 33 miRNAs and 2,791 mRNAs were significantly dysregulated between the two groups. Among the 2,791 mRNAs, 911 overlapped with putative miRNA target genes. A pathway analysis showed that the MAPK pathway was consistently enriched in the chordoma tissue and that miR-149-3p, miR-663a, miR-1908, miR-2861 and miR-3185 likely play important roles in the regulation of MAPK pathways. Furthermore, the Notch signaling pathway and the loss of the calcification or ossification capacity of the notochord may also be involved in chordoma pathogenesis.
Conclusion: This study provides an integrated dataset of the miRNA and mRNA profiles in chordomas, and the results demonstrate that not only the MAPK pathway and its related miRNAs but also the Notch pathway may be involved in chordoma development. The occurrence of chordoma may be associated with dysfunctional calcification or ossification of the notochord.
Conflict of interest statement
Figures




References
-
- Walcott BP, Nahed BV, Mohyeldin A, Coumans JV, Kahle KT, et al. (2012) Chordoma: current concepts, management, and future directions. Lancet Oncol 13: e69–76. - PubMed
-
- Scheil S, Bruderlein S, Liehr T, Starke H, Herms J, et al. (2001) Genome-wide analysis of sixteen chordomas by comparative genomic hybridization and cytogenetics of the first human chordoma cell line, U-CH1. Genes Chromosomes Cancer 32: 203–211. - PubMed
-
- Chugh R, Tawbi H, Lucas DR, Biermann JS, Schuetze SM, et al. (2007) Chordoma: the nonsarcoma primary bone tumor. Oncologist 12: 1344–1350. - PubMed
-
- Deniz ML, Kilic T, Almaata I, Kurtkaya O, Sav A, et al.. (2002) Expression of growth factors and structural proteins in chordomas: basic fibroblast growth factor, transforming growth factor alpha, and fibronectin are correlated with recurrence. Neurosurgery 51: 753–760; discussion 760. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

