SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature
- PMID: 23826184
- PMCID: PMC3691135
- DOI: 10.1371/journal.pone.0067003
SLE peripheral blood B cell, T cell and myeloid cell transcriptomes display unique profiles and each subset contributes to the interferon signature
Abstract
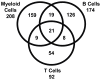
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease that is characterized by defective immune tolerance combined with immune cell hyperactivity resulting in the production of pathogenic autoantibodies. Previous gene expression studies employing whole blood or peripheral blood mononuclear cells (PBMC) have demonstrated that a majority of patients with active disease have increased expression of type I interferon (IFN) inducible transcripts known as the IFN signature. The goal of the current study was to assess the gene expression profiles of isolated leukocyte subsets obtained from SLE patients. Subsets including CD19(+) B lymphocytes, CD3(+)CD4(+) T lymphocytes and CD33(+) myeloid cells were simultaneously sorted from PBMC. The SLE transcriptomes were assessed for differentially expressed genes as compared to healthy controls. SLE CD33(+) myeloid cells exhibited the greatest number of differentially expressed genes at 208 transcripts, SLE B cells expressed 174 transcripts and SLE CD3(+)CD4(+) T cells expressed 92 transcripts. Only 4.4% (21) of the 474 total transcripts, many associated with the IFN signature, were shared by all three subsets. Transcriptional profiles translated into increased protein expression for CD38, CD63, CD107a and CD169. Moreover, these studies demonstrated that both SLE lymphoid and myeloid subsets expressed elevated transcripts for cytosolic RNA and DNA sensors and downstream effectors mediating IFN and cytokine production. Prolonged upregulation of nucleic acid sensing pathways could modulate immune effector functions and initiate or contribute to the systemic inflammation observed in SLE.
Conflict of interest statement
Figures






References
-
- Crow MK (2008) Collaboration, genetic associations, and lupus erythematosus. N Engl J Med 358: 956–961. - PubMed
-
- Petri M (1995) Clinical features of systemic lupus erythematosus. Curr Opin Rheumatol 7: 395–401. - PubMed
-
- Arbuckle MR, McClain MT, Rubertone MV, Scofield RH, Dennis GJ, et al. (2003) Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med 349: 1526–1533. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

