Sequencing of the Hepatitis C Virus: A Systematic Review
- PMID: 23826196
- PMCID: PMC3694929
- DOI: 10.1371/journal.pone.0067073
Sequencing of the Hepatitis C Virus: A Systematic Review
Abstract
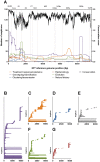
Since the identification of hepatitis C virus (HCV), viral sequencing has been important in understanding HCV classification, epidemiology, evolution, transmission clustering, treatment response and natural history. The length and diversity of the HCV genome has resulted in analysis of certain regions of the virus, however there has been little standardisation of protocols. This systematic review was undertaken to map the location and frequency of sequencing on the HCV genome in peer reviewed publications, with the aim to produce a database of sequencing primers and amplicons to inform future research. Medline and Scopus databases were searched for English language publications based on keyword/MeSH terms related to sequence analysis (9 terms) or HCV (3 terms), plus "primer" as a general search term. Exclusion criteria included non-HCV research, review articles, duplicate records, and incomplete description of HCV sequencing methods. The PCR primer locations of accepted publications were noted, and purpose of sequencing was determined. A total of 450 studies were accepted from the 2099 identified, with 629 HCV sequencing amplicons identified and mapped on the HCV genome. The most commonly sequenced region was the HVR-1 region, often utilised for studies of natural history, clustering/transmission, evolution and treatment response. Studies related to genotyping/classification or epidemiology of HCV genotype generally targeted the 5'UTR, Core and NS5B regions, while treatment response/resistance was assessed mainly in the NS3-NS5B region with emphasis on the Interferon sensitivity determining region (ISDR) region of NS5A. While the sequencing of HCV is generally constricted to certain regions of the HCV genome there is little consistency in the positioning of sequencing primers, with the exception of a few highly referenced manuscripts. This study demonstrates the heterogeneity of HCV sequencing, providing a comprehensive database of previously published primer sets to be utilised in future sequencing studies.
Conflict of interest statement
Figures
References
-
- Saldanha J, Lelie N, Heath A, Grp WHOCS (1999) Establishment of the first international standard for nucleic acid amplification technology (NAT) assays for HCV RNA. Vox Sanguinis 76: 149–158. - PubMed
-
- Simmonds P, Alberti A, Alter HJ, Bonino F, Bradley DW, et al. (1994) A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19: 1321–1324. - PubMed
-
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, et al. (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962–973. - PubMed
-
- Stumpf MPH, Pybus OG (2002) Genetic diversity and models of viral evolution for the hepatitis C virus. Fems Microbiology Letters 214: 143–152. - PubMed
-
- Pawlotsky JM (2006) Hepatitis C virus population dynamics during infection. Quasispecies: Concept and Implications for Virology 299: 261–284. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

