Comparative Genomics Reveal That Host-Innate Immune Responses Influence the Clinical Prevalence of Legionella pneumophila Serogroups
- PMID: 23826259
- PMCID: PMC3694923
- DOI: 10.1371/journal.pone.0067298
Comparative Genomics Reveal That Host-Innate Immune Responses Influence the Clinical Prevalence of Legionella pneumophila Serogroups
Abstract
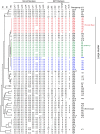
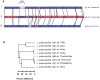
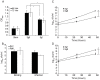
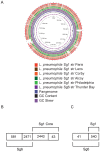
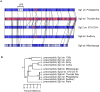
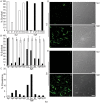
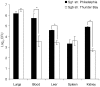
Legionella pneumophila is the primary etiologic agent of legionellosis, a potentially fatal respiratory illness. Amongst the sixteen described L. pneumophila serogroups, a majority of the clinical infections diagnosed using standard methods are serogroup 1 (Sg1). This high clinical prevalence of Sg1 is hypothesized to be linked to environmental specific advantages and/or to increased virulence of strains belonging to Sg1. The genetic determinants for this prevalence remain unknown primarily due to the limited genomic information available for non-Sg1 clinical strains. Through a systematic attempt to culture Legionella from patient respiratory samples, we have previously reported that 34% of all culture confirmed legionellosis cases in Ontario (n = 351) are caused by non-Sg1 Legionella. Phylogenetic analysis combining multiple-locus variable number tandem repeat analysis and sequence based typing profiles of all non-Sg1 identified that L. pneumophila clinical strains (n = 73) belonging to the two most prevalent molecular types were Sg6. We conducted whole genome sequencing of two strains representative of these sequence types and one distant neighbour. Comparative genomics of the three L. pneumophila Sg6 genomes reported here with published L. pneumophila serogroup 1 genomes identified genetic differences in the O-antigen biosynthetic cluster. Comparative optical mapping analysis between Sg6 and Sg1 further corroborated this finding. We confirmed an altered O-antigen profile of Sg6, and tested its possible effects on growth and replication in in vitro biological models and experimental murine infections. Our data indicates that while clinical Sg1 might not be better suited than Sg6 in colonizing environmental niches, increased bloodstream dissemination through resistance to the alternative pathway of complement mediated killing in the human host may explain its higher prevalence.
Conflict of interest statement
Figures

Similar articles
-
Prevalence of Infection-Competent Serogroup 6 Legionella pneumophila within Premise Plumbing in Southeast Michigan.mBio. 2018 Feb 6;9(1):e00016-18. doi: 10.1128/mBio.00016-18. mBio. 2018. PMID: 29437918 Free PMC article.
-
Multigenome analysis identifies a worldwide distributed epidemic Legionella pneumophila clone that emerged within a highly diverse species.Genome Res. 2008 Mar;18(3):431-41. doi: 10.1101/gr.7229808. Epub 2008 Feb 6. Genome Res. 2008. PMID: 18256241 Free PMC article.
-
Legionellae isolated from clinical and environmental samples in Spain (1983-90): monoclonal typing of Legionella pneumophila serogroup 1 isolates.Epidemiol Infect. 1992 Jun;108(3):397-402. doi: 10.1017/s0950268800049906. Epidemiol Infect. 1992. PMID: 1601074 Free PMC article.
-
Legionella pneumophila: population genetics, phylogeny and genomics.Infect Genet Evol. 2009 Sep;9(5):727-39. doi: 10.1016/j.meegid.2009.05.004. Epub 2009 May 18. Infect Genet Evol. 2009. PMID: 19450709 Review.
-
Molecular pathogenesis of infections caused by Legionella pneumophila.Clin Microbiol Rev. 2010 Apr;23(2):274-98. doi: 10.1128/CMR.00052-09. Clin Microbiol Rev. 2010. PMID: 20375353 Free PMC article. Review.
Cited by
-
Current and emerging Legionella diagnostics for laboratory and outbreak investigations.Clin Microbiol Rev. 2015 Jan;28(1):95-133. doi: 10.1128/CMR.00029-14. Clin Microbiol Rev. 2015. PMID: 25567224 Free PMC article. Review.
-
Epidemiological Investigation of Legionella pneumophila Serogroup 2 to 14 Isolates from Water Samples by Amplified Fragment Length Polymorphism and Sequence-Based Typing and Detection of Virulence Traits.Appl Environ Microbiol. 2016 Sep 30;82(20):6102-6108. doi: 10.1128/AEM.01672-16. Print 2016 Oct 15. Appl Environ Microbiol. 2016. PMID: 27496776 Free PMC article.
-
Genomic heterogeneity differentiates clinical and environmental subgroups of Legionella pneumophila sequence type 1.PLoS One. 2018 Oct 18;13(10):e0206110. doi: 10.1371/journal.pone.0206110. eCollection 2018. PLoS One. 2018. PMID: 30335848 Free PMC article.
-
The Legionella pneumophila collagen-like protein mediates sedimentation, autoaggregation, and pathogen-phagocyte interactions.Appl Environ Microbiol. 2014 Feb;80(4):1441-54. doi: 10.1128/AEM.03254-13. Epub 2013 Dec 13. Appl Environ Microbiol. 2014. PMID: 24334670 Free PMC article.
-
Comparative Genomics of Legionella pneumophila Isolates from the West Bank and Germany Support Molecular Epidemiology of Legionnaires' Disease.Microorganisms. 2023 Feb 10;11(2):449. doi: 10.3390/microorganisms11020449. Microorganisms. 2023. PMID: 36838414 Free PMC article.
References
-
- Fraser DW, Tsai TR, Orenstein W, Parkin WE, Beecham HJ, et al. (1977) Legionnaires' disease: description of an epidemic of pneumonia. N Engl J Med 297: 1189–1197. - PubMed
-
- Sanford JP (1979) Legionnaires' disease: one person's perspective. Ann Intern Med 90: 699–703. - PubMed
-
- McDade JE, Shepard CC, Fraser DW, Tsai TR, Redus MA, et al. (1977) Legionnaires' disease: isolation of a bacterium and demonstration of its role in other respiratory disease. N Engl J Med 297: 1197–1203. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

