Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan
- PMID: 23826282
- PMCID: PMC3695075
- DOI: 10.1371/journal.pone.0067378
Continuous Aging of the Human DNA Methylome Throughout the Human Lifespan
Abstract
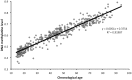
DNA methylation plays an important role in development of disease and the process of aging. In this study we examine DNA methylation at 476,366 sites throughout the genome of white blood cells from a population cohort (N = 421) ranging in age from 14 to 94 years old. Age affects DNA methylation at almost one third (29%) of the sites (Bonferroni adjusted P-value <0.05), of which 60.5% becomes hypomethylated and 39.5% hypermethylated with increasing age. DNA methylation sites that are located within CpG islands (CGIs) more often become hypermethylated compared to sites outside an island. CpG sites in promoters are more unaffected by age, whereas sites in enhancers more often becomes hypo- or hypermethylated. Hypermethylated sites are overrepresented among genes that are involved in DNA binding, transcription regulation, processes of anatomical structure and developmental process and cortex neuron differentiation (P-value down to P = 9.14*10(-67)). By contrast, hypomethylated sites are not strongly overrepresented among any biological function or process. Our results indicate that the 23% of the variation in DNA methylation is attributed chronological age, and that hypermethylation is more site-specific than hypomethylation. It appears that the change in DNA methylation partly overlap with regions that change histone modifications with age, indicating an interaction between the two major epigenetic mechanisms. Epigenetic modifications and change in gene expression over time most likely reflects the natural process of aging and variation between individuals might contribute to the development of age-related phenotypes and diseases such as type II diabetes, autoimmune and cardiovascular disease.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

