Structural basis of carotenoid cleavage: from bacteria to mammals
- PMID: 23827316
- PMCID: PMC3818509
- DOI: 10.1016/j.abb.2013.06.012
Structural basis of carotenoid cleavage: from bacteria to mammals
Abstract
Carotenoids and their metabolic derivatives serve critical functions in both prokaryotic and eukaryotic cells, including pigmentation, photoprotection and photosynthesis as well as cell signaling. These organic compounds are also important for visual function in vertebrate and non-vertebrate organisms. Enzymatic transformations of carotenoids to various apocarotenoid products are catalyzed by a family of evolutionarily conserved, non-heme iron-containing enzymes named carotenoid cleavage oxygenases (CCOs). Studies have revealed that CCOs are critically involved in carotenoid homeostasis and essential for the health of organisms including humans. These enzymes typically display a high degree of regio- and stereo-selectivity, acting on specific positions of the polyene backbone located in their substrates. By oxidatively cleaving and/or isomerizing specific double bonds, CCOs generate a variety of apocarotenoid isomer products. Recent structural studies have helped illuminate the mechanisms by which CCOs mobilize their lipophilic substrates from biological membranes to perform their characteristic double bond cleavage and/or isomerization reactions. In this review, we aim to integrate structural and biochemical information about CCOs to provide insights into their catalytic mechanisms.
Keywords: ACO; Apocarotenoid; Carotenoid; Carotenoid oxygenase; RPE65; VP14.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
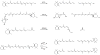

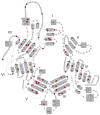
 ) and their fixating Glu residues (
) and their fixating Glu residues (
 ) are labeled. Dots mark every tenth residue. The sequences were aligned with with T-coffee [75] and the figure was generated with ESPript [76].
) are labeled. Dots mark every tenth residue. The sequences were aligned with with T-coffee [75] and the figure was generated with ESPript [76].
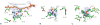

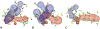

References
-
- Walter MH, Strack D. Natural product reports. 2011;28:663–692. - PubMed
-
- Blount JD. Archives of biochemistry and biophysics. 2004;430:10–15. - PubMed
-
- Fraser NJ, Hashimoto H, Cogdell RJ. Photosynthesis research. 2001;70:249–256. - PubMed
-
- Stahl W, Sies H. The American journal of clinical nutrition. 2012;96:1179S–1184S. - PubMed
-
- Carranco Jauregui ME, Calvo Carrillo L, Mde, Romo FP. Archivos latinoamericanos de nutricion. 2011;61:233–241. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

