lincRNAs: genomics, evolution, and mechanisms
- PMID: 23827673
- PMCID: PMC3924787
- DOI: 10.1016/j.cell.2013.06.020
lincRNAs: genomics, evolution, and mechanisms
Abstract
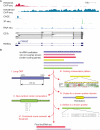
Long intervening noncoding RNAs (lincRNAs) are transcribed from thousands of loci in mammalian genomes and might play widespread roles in gene regulation and other cellular processes. This Review outlines the emerging understanding of lincRNAs in vertebrate animals, with emphases on how they are being identified and current conclusions and questions regarding their genomics, evolution and mechanisms of action.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

