Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking
- PMID: 23827676
- PMCID: PMC3756801
- DOI: 10.1016/j.cell.2013.05.060
Presynaptic neurexin-3 alternative splicing trans-synaptically controls postsynaptic AMPA receptor trafficking
Abstract
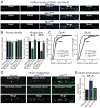
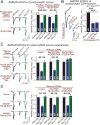
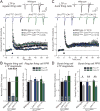
Neurexins are essential presynaptic cell adhesion molecules that are linked to schizophrenia and autism and are subject to extensive alternative splicing. Here, we used a genetic approach to test the physiological significance of neurexin alternative splicing. We generated knockin mice in which alternatively spliced sequence #4 (SS4) of neuexin-3 is constitutively included but can be selectively excised by cre-recombination. SS4 of neurexin-3 was chosen because it is highly regulated and controls neurexin binding to neuroligins, LRRTMs, and other ligands. Unexpectedly, constitutive inclusion of SS4 in presynaptic neurexin-3 decreased postsynaptic AMPA, but not NMDA receptor levels, and enhanced postsynaptic AMPA receptor endocytosis. Moreover, constitutive inclusion of SS4 in presynaptic neurexin-3 abrogated postsynaptic AMPA receptor recruitment during NMDA receptor-dependent LTP. These phenotypes were fully rescued by constitutive excision of SS4 in neurexin-3. Thus, alternative splicing of presynaptic neurexin-3 controls postsynaptic AMPA receptor trafficking, revealing an unanticipated alternative splicing mechanism for trans-synaptic regulation of synaptic strength and long-term plasticity.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice code for trans-synaptic cell adhesion mediated by binding of neuroligin 1 to α- and β-neurexins. Neuron. 2005;48:229–236. - PubMed
-
- Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

