Observing a DNA polymerase choose right from wrong
- PMID: 23827680
- PMCID: PMC3924593
- DOI: 10.1016/j.cell.2013.05.048
Observing a DNA polymerase choose right from wrong
Abstract
DNA polymerase (pol) β is a model polymerase involved in gap-filling DNA synthesis utilizing two metals to facilitate nucleotidyl transfer. Previous structural studies have trapped catalytic intermediates by utilizing substrate analogs (dideoxy-terminated primer or nonhydrolysable incoming nucleotide). To identify additional intermediates during catalysis, we now employ natural substrates (correct and incorrect nucleotides) and follow product formation in real time with 15 different crystal structures. We are able to observe molecular adjustments at the active site that hasten correct nucleotide insertion and deter incorrect insertion not appreciated previously. A third metal binding site is transiently formed during correct, but not incorrect, nucleotide insertion. Additionally, long incubations indicate that pyrophosphate more easily dissociates after incorrect, compared to correct, nucleotide insertion. This appears to be coupled to subdomain repositioning that is required for catalytic activation/deactivation. The structures provide insights into a fundamental chemical reaction that impacts polymerase fidelity and genome stability.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




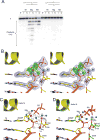

Comment in
-
Watching a DNA polymerase in action.Cell Cycle. 2014;13(5):691-2. doi: 10.4161/cc.27789. Epub 2014 Jan 14. Cell Cycle. 2014. PMID: 24424116 Free PMC article. No abstract available.
References
-
- Beard WA, Shock DD, Yang XP, DeLauder SF, Wilson SH. Loss of DNA polymerase β stacking interactions with templating purines, but not pyrimidines, alters catalytic efficiency and fidelity. J Biol Chem. 2002;277:8235–8242. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

