Identification of active regulatory regions from DNA methylation data
- PMID: 23828043
- PMCID: PMC3763559
- DOI: 10.1093/nar/gkt599
Identification of active regulatory regions from DNA methylation data
Abstract
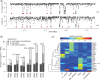
We have recently shown that transcription factor binding leads to defined reduction in DNA methylation, allowing for the identification of active regulatory regions from high-resolution methylomes. Here, we present MethylSeekR, a computational tool to accurately identify such footprints from bisulfite-sequencing data. Applying our method to a large number of published human methylomes, we demonstrate its broad applicability and generalize our previous findings from a neuronal differentiation system to many cell types and tissues. MethylSeekR is available as an R package at www.bioconductor.org.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

