Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a)
- PMID: 23828779
- PMCID: PMC3770094
- DOI: 10.1194/jlr.M040733
Determinants of binding of oxidized phospholipids on apolipoprotein (a) and lipoprotein (a)
Abstract
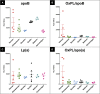
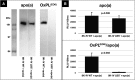
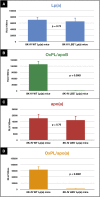
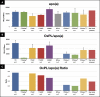
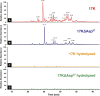
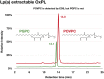
Oxidized phospholipids (OxPLs) are present on apolipoprotein (a) [apo(a)] and lipoprotein (a) [Lp(a)] but the determinants influencing their binding are not known. The presence of OxPLs on apo(a)/Lp(a) was evaluated in plasma from healthy humans, apes, monkeys, apo(a)/Lp(a) transgenic mice, lysine binding site (LBS) mutant apo(a)/Lp(a) mice with Asp(55/57)→Ala(55/57) substitution of kringle (K)IV10)], and a variety of recombinant apo(a) [r-apo(a)] constructs. Using antibody E06, which binds the phosphocholine (PC) headgroup of OxPLs, Western and ELISA formats revealed that OxPLs were only present in apo(a) with an intact KIV10 LBS. Lipid extracts of purified human Lp(a) contained both E06- and nonE06-detectable OxPLs by tandem liquid chromatography-mass spectrometry (LC-MS/MS). Trypsin digestion of 17K r-apo(a) showed PC-containing OxPLs covalently bound to apo(a) fragments by LC-MS/MS that could be saponified by ammonium hydroxide. Interestingly, PC-containing OxPLs were also present in 17K r-apo(a) with Asp(57)→Ala(57) substitution in KIV10 that lacked E06 immunoreactivity. In conclusion, E06- and nonE06-detectable OxPLs are present in the lipid phase of Lp(a) and covalently bound to apo(a). E06 immunoreactivity, reflecting pro-inflammatory OxPLs accessible to the immune system, is strongly influenced by KIV10 LBS and is unique to human apo(a), which may explain Lp(a)'s pro-atherogenic potential.
Keywords: kringles; lipoproteins; plasminogen.
Figures



References
-
- Dubé J. B., Boffa M. B., Hegele R. A., Koschinsky M. L. 2012. Lipoprotein(a): more interesting than ever after 50 years. Curr. Opin. Lipidol. 23: 133–140 - PubMed
-
- Kronenberg F., Utermann G. 2013. Lipoprotein(a): resurrected by genetics. J. Intern. Med. 273: 6–30 - PubMed
-
- McLean J. W., Tomlinson J. E., Kuang W. J., Eaton D. L., Chen E. Y., Fless G. M., Scanu A. M., Lawn R. M. 1987. cDNA sequence of human apolipoprotein(a) is homologous to plasminogen. Nature. 330: 132–137 - PubMed
-
- Lawn R. M., Boonmark N. W., Schwartz K., Lindahl G. E., Wade D. P., Byrne C. D., Fong K. J., Meer K., Patthy L. 1995. The recurring evolution of lipoprotein(a). Insights from cloning of hedgehog apolipoprotein(a). J. Biol. Chem. 270: 24004–24009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

