A common 56-kilobase deletion in a primate-specific segmental duplication creates a novel butyrophilin-like protein
- PMID: 23829304
- PMCID: PMC3729544
- DOI: 10.1186/1471-2156-14-61
A common 56-kilobase deletion in a primate-specific segmental duplication creates a novel butyrophilin-like protein
Abstract
Background: The Butyrophilin-like (BTNL) proteins are likely to play an important role in inflammation and immune response. Like the B7 protein family, many human and murine BTNL members have been shown to control T lymphocytes response, and polymorphisms in human BTNL2 have been linked to several inflammatory diseases, such as pulmonary sarcoidosis, inflammatory bowel disease and neonatal lupus.
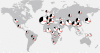
Results: In this study we provide a comprehensive population, genomic and transcriptomic analysis of a 56-kb deletion copy number variant (CNV), located within two segmental duplications of two genes belonging to the BTNL family, namely BTNL8 and BTNL3. We confirm the presence of a novel BTNL8*3 fusion-protein product, and show an influence of the deletion variant on the expression level of several genes involved in immune function, including BTNL9, another member of the same family. Moreover, by genotyping HapMap and human diversity panel (HGDP) samples, we demonstrate a clear difference in the stratification of the BTNL8_BTNL3-del allele frequency between major continental human populations.
Conclusion: Despite tremendous progress in the field of structural variation, rather few CNVs have been functionally characterized so far. Here, we show clear functional consequences of a new deletion CNV (BTNL8_BTNL3-del) with potentially important implication in the human immune system and in inflammatory and proliferative disorders. In addition, the marked population differences found of BTNL8_BTNL3-del frequencies suggest that this deletion CNV might have evolved under positive selection due to environmental conditions in some populations, with potential phenotypic consequences.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

