An untargeted metabolomic workflow to improve structural characterization of metabolites
- PMID: 23829391
- PMCID: PMC3983953
- DOI: 10.1021/ac400751j
An untargeted metabolomic workflow to improve structural characterization of metabolites
Abstract
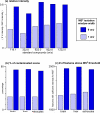
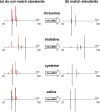
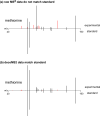
Mass spectrometry-based metabolomics relies on MS(2) data for structural characterization of metabolites. To obtain the high-quality MS(2) data necessary to support metabolite identifications, ions of interest must be purely isolated for fragmentation. Here, we show that metabolomic MS(2) data are frequently characterized by contaminating ions that prevent structural identification. Although using narrow-isolation windows can minimize contaminating MS(2) fragments, even narrow windows are not always selective enough, and they can complicate data analysis by removing isotopic patterns from MS(2) spectra. Moreover, narrow windows can significantly reduce sensitivity. In this work, we introduce a novel, two-part approach for performing metabolomic identifications that addresses these issues. First, we collect MS(2) scans with less stringent isolation settings to obtain improved sensitivity at the expense of specificity. Then, by evaluating MS(2) fragment intensities as a function of retention time and precursor mass targeted for MS(2) analysis, we obtain deconvolved MS(2) spectra that are consistent with pure standards and can therefore be used for metabolite identification. The value of our approach is highlighted with metabolic extracts from brain, liver, astrocytes, as well as nerve tissue, and performance is evaluated by using pure metabolite standards in combination with simulations based on raw MS(2) data from the METLIN metabolite database. A R package implementing the algorithms used in our workflow is available on our laboratory website ( http://pattilab.wustl.edu/decoms2.php ).
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

