Role of small nuclear RNAs in eukaryotic gene expression
- PMID: 23829528
- PMCID: PMC11246792
- DOI: 10.1042/bse0540079
Role of small nuclear RNAs in eukaryotic gene expression
Abstract
Eukaryotic cells contain small, highly abundant, nuclear-localized non-coding RNAs [snRNAs (small nuclear RNAs)] which play important roles in splicing of introns from primary genomic transcripts. Through a combination of RNA-RNA and RNA-protein interactions, two of the snRNPs, U1 and U2, recognize the splice sites and the branch site of introns. A complex remodelling of RNA-RNA and protein-based interactions follows, resulting in the assembly of catalytically competent spliceosomes, in which the snRNAs and their bound proteins play central roles. This process involves formation of extensive base-pairing interactions between U2 and U6, U6 and the 5' splice site, and U5 and the exonic sequences immediately adjacent to the 5' and 3' splice sites. Thus RNA-RNA interactions involving U2, U5 and U6 help position the reacting groups of the first and second steps of splicing. In addition, U6 is also thought to participate in formation of the spliceosomal active site. Furthermore, emerging evidence suggests additional roles for snRNAs in regulation of various aspects of RNA biogenesis, from transcription to polyadenylation and RNA stability. These snRNP-mediated regulatory roles probably serve to ensure the co-ordination of the different processes involved in biogenesis of RNAs and point to the central importance of snRNAs in eukaryotic gene expression.
Figures


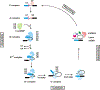
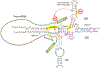

References
-
- Will CL, Luhrmann R: Spliceosome structure and function. In The RNA World. Edited by Gesteland RF, Cech TR, Atkins JF. Cold Spring Harbor Laboratory Press; 2006:369–400.
-
- Roy SW, Irimia M (2009)Splicing in the eukaryotic ancestor: form, function and dysfunction. Trends Ecol. Evol. (Amst.) 24:447–455. - PubMed
-
- Rodríguez-Trelles F, Tarrío R, Ayala FJ (2006) Origins and evolution of spliceosomal introns. Annu. Rev. Genet 40:47–76. - PubMed
-
- Valadkhan S (2010) Role of the snRNAs in spliceosomal active site. RNA Biol, 7:345–353. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

