Rank-based genome-wide analysis reveals the association of ryanodine receptor-2 gene variants with childhood asthma among human populations
- PMID: 23829686
- PMCID: PMC3708719
- DOI: 10.1186/1479-7364-7-16
Rank-based genome-wide analysis reveals the association of ryanodine receptor-2 gene variants with childhood asthma among human populations
Abstract
Background: The standard approach to determine unique or shared genetic factors across populations is to identify risk alleles in one population and investigate replication in others. However, since populations differ in DNA sequence information, allele frequencies, effect sizes, and linkage disequilibrium patterns, SNP association using a uniform stringent threshold on p values may not be reproducible across populations. Here, we developed rank-based methods to investigate shared or population-specific loci and pathways for childhood asthma across individuals of diverse ancestry. We performed genome-wide association studies on 859,790 SNPs genotyped in 527 affected offspring trios of European, African, and Hispanic ancestry using publically available asthma database in the Genotypes and Phenotypes database.
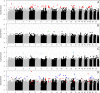
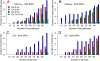
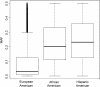
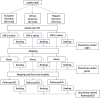
Results: Rank-based analyses showed that there are shared genetic factors for asthma across populations, more at the gene and pathway levels than at the SNP level. Although the top 1,000 SNPs were not shared, 11 genes (RYR2, PDE4D, CSMD1, CDH13, ROBO2, RBFOX1, PTPRD, NPAS3, PDE1C, SEMA5A, and CTNNA2) mapped by these SNPs were shared across populations. Ryanodine receptor 2 (RYR2, a statin response-related gene) showed the strongest association in European (p value=2.55×10(-7)) and was replicated in African (2.57×10(-4)) and Hispanic (1.18 × 10(-3)) Americans. Imputation analyses based on the 1000 Genomes Project uncovered additional RYR2 variants associated with asthma. Network and functional ontology analyses revealed that RYR2 is an integral part of dermatological or allergic disorder biological networks, specifically in the functional classes involving inflammatory, eosinophilic, and respiratory diseases.
Conclusion: Our rank-based genome-wide analysis revealed for the first time an association of RYR2 variants with asthma and replicated previously discovered PDE4D asthma gene across human populations. The replication of top-ranked asthma genes across populations suggests that such loci are less likely to be false positives and could indicate true associations. Variants that are associated with asthma across populations could be used to identify individuals who are at high risk for asthma regardless of genetic ancestry.
Figures



References
-
- Akinbami LJ, Liu X, Pastor PN, Reuben CA. Data from the national health interview survey, 1998–2009. NCHS Data Brief. 2011. pp. 1–8. - PubMed
-
- Moorman JE, Rudd RA, Johnson CA, King M, Minor P, Bailey C, Scalia MR, Akinbami LJ. National surveillance for asthma–United States, 1980–2004. MMWR Surveill Summ. 2007;56(8):1–54. - PubMed
-
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

