Breaking and joining single-stranded DNA: the HUH endonuclease superfamily
- PMID: 23832240
- PMCID: PMC6493337
- DOI: 10.1038/nrmicro3067
Breaking and joining single-stranded DNA: the HUH endonuclease superfamily
Abstract
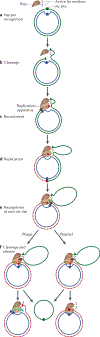
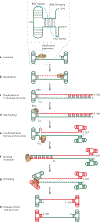
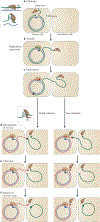
HUH endonucleases are numerous and widespread in all three domains of life. The major function of these enzymes is processing a range of mobile genetic elements by catalysing cleavage and rejoining of single-stranded DNA using an active-site Tyr residue to make a transient 5'-phosphotyrosine bond with the DNA substrate. These enzymes have a key role in rolling-circle replication of plasmids and bacteriophages, in plasmid transfer, in the replication of several eukaryotic viruses and in various types of transposition. They have also been appropriated for cellular processes such as intron homing and the processing of bacterial repeated extragenic palindromes. Here, we provide an overview of these fascinating enzymes and their functions, using well-characterized examples of Rep proteins, relaxases and transposases, and we explore the molecular mechanisms used in their diverse activities.
Conflict of interest statement
Competing interests statement
The authors declare no competing financial interests.
Figures
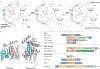
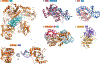

References
-
- Kornberg A & Baker TA DNA Replication 2nd edn (Freeman, 1992).
-
-
Koonin EV & Ilyina TV Computer-assisted dissection of rolling circle DNA replication. Biosystems 30, 241–268 (1993).
References 2 and 3 are the first bioinformatic analyses of HUH endonucleases.
-
-
- Garcillan-Barcia MP, Bernales I, Mendiola MV & De la Cruz F in Mobile DNA Vol. II (eds Craig NL, Craigie R, Gellert M, & Lambowitz A) 891–904 (ASM Press, 2002).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

