Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods?
- PMID: 23832846
- PMCID: PMC3874840
- DOI: 10.1002/bies.201300040
Segmental folding of chromosomes: a basis for structural and regulatory chromosomal neighborhoods?
Abstract
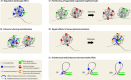
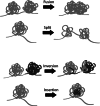
We discuss here a series of testable hypotheses concerning the role of chromosome folding into topologically associating domains (TADs). Several lines of evidence suggest that segmental packaging of chromosomal neighborhoods may underlie features of chromatin that span large domains, such as heterochromatin blocks, association with the nuclear lamina and replication timing. By defining which DNA elements preferentially contact each other, the segmentation of chromosomes into TADs may also underlie many properties of long-range transcriptional regulation. Several observations suggest that TADs can indeed provide a structural basis to regulatory landscapes, by controlling enhancer sharing and allocation. We also discuss how TADs may shape the evolution of chromosomes, by causing maintenance of synteny over large chromosomal segments. Finally we suggest a series of experiments to challenge these ideas and provide concrete examples illustrating how they could be practically applied.
Keywords: chromatin domains; chromatin folding; chromosome conformation capture (3C); long-range transcriptional regulation; regulatory landscapes; topologically associating chromosome domains.
© 2013 The Authors. Bioessays published by WILEY Periodicals, Inc.
Figures



References
-
- Dekker J, Klecker N. Capturing chromosome conformation. Science. 2002;295:1306–11. - PubMed
-
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, et al. Looping and interaction between hypersensitive sites in the active [beta]-globin locus. Mol Cell. 2002;10:1453–65. - PubMed
-
- Simonis M, Klous P, Splinter E, Moshkin Y, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on-chip (4C) Nat Genet. 2006;38:1348–54. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

