Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation
- PMID: 23834507
- PMCID: PMC3716973
- DOI: 10.1186/1471-2164-14-460
Transcriptional profiling of sweetpotato (Ipomoea batatas) roots indicates down-regulation of lignin biosynthesis and up-regulation of starch biosynthesis at an early stage of storage root formation
Abstract
Background: The number of fibrous roots that develop into storage roots determines sweetpotato yield. The aim of the present study was to identify the molecular mechanisms involved in the initiation of storage root formation, by performing a detailed transcriptomic analysis of initiating storage roots using next-generation sequencing platforms. A two-step approach was undertaken: (1) generating a database for the sweetpotato root transcriptome using 454-Roche sequencing of a cDNA library created from pooled samples of two root types: fibrous and initiating storage roots; (2) comparing the expression profiles of initiating storage roots and fibrous roots, using the Illumina Genome Analyzer to sequence cDNA libraries of the two root types and map the data onto the root transcriptome database.
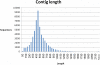
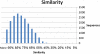
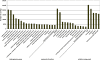
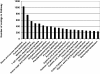
Results: Use of the 454-Roche platform generated a total of 524,607 reads, 85.6% of which were clustered into 55,296 contigs that matched 40,278 known genes. The reads, generated by the Illumina Genome Analyzer, were found to map to 31,284 contigs out of the 55,296 contigs serving as the database. A total of 8,353 contigs were found to exhibit differential expression between the two root types (at least 2.5-fold change). The Illumina-based differential expression results were validated for nine putative genes using quantitative real-time PCR. The differential expression profiles indicated down-regulation of classical root functions, such as transport, as well as down-regulation of lignin biosynthesis in initiating storage roots, and up-regulation of carbohydrate metabolism and starch biosynthesis. In addition, data indicated delicate control of regulators of meristematic tissue identity and maintenance, associated with the initiation of storage root formation.
Conclusions: This study adds a valuable resource of sweetpotato root transcript sequences to available data, facilitating the identification of genes of interest. This resource enabled us to identify genes that are involved in the earliest stage of storage root formation, highlighting the reduction in carbon flow toward phenylpropanoid biosynthesis and its delivery into carbohydrate metabolism and starch biosynthesis, as major events involved in storage root initiation. The novel transcripts related to storage root initiation identified in this study provide a starting point for further investigation into the molecular mechanisms underlying this process.
Figures







References
-
- FAOSTAT Data. [ http://faostat3.fao.org/home/index.html]
-
- Woolfe J. Sweetpotato, an Untapped Food Source. Cambridge: Cambridge University Press; 1992.
-
- The Nutrition Action Health Letter from the Center for Science in the Public Interest. USA; [ http://www.cspinet.org/nah/]
-
- Togari Y. A study of tuberous root formation in sweet potato. Bull Natl Agric Exp Statn Tokyo. 1950;68:1–96.
-
- Wilson LA, Lowe SB. The anatomy of the root system in West Indian sweet potato [Ipomoea batatas (L.) Lam.] cultivars. Ann Bot (Lond) 1973;37:633–643.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

