Deletion of creB in Aspergillus oryzae increases secreted hydrolytic enzyme activity
- PMID: 23835170
- PMCID: PMC3754153
- DOI: 10.1128/AEM.01406-13
Deletion of creB in Aspergillus oryzae increases secreted hydrolytic enzyme activity
Abstract
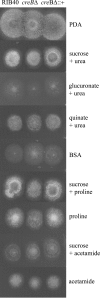
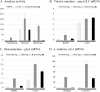
Aspergillus oryzae has been used in the food and beverage industry for centuries, and industrial strains have been produced by multiple rounds of selection. Targeted gene deletion technology is particularly useful for strain improvement in such strains, particularly when they do not have a well-characterized meiotic cycle. Phenotypes of an Aspergillus nidulans strain null for the CreB deubiquitinating enzyme include effects on growth and repression, including increased activity levels of various enzymes. We show that Aspergillus oryzae contains a functional homologue of the CreB deubiquitinating enzyme and that a null strain shows increased activity levels of industrially important secreted enzymes, including cellulases, xylanases, amylases, and proteases, as well as alleviated inhibition of spore germination on glucose medium. Reverse transcription-quantitative PCR (RT-qPCR) analysis showed that the increased levels of enzyme activity in both Aspergillus nidulans and Aspergillus oryzae are mirrored at the transcript level, indicating transcriptional regulation. We report that Aspergillus oryzae DAR3699, originally isolated from soy fermentation, has a similar phenotype to that of a creB deletion mutant of the RIB40 strain, and it contains a mutation in the creB gene. Collectively, the results for Aspergillus oryzae, Aspergillus nidulans, Trichoderma reesei, and Penicillium decumbens show that deletion of creB may be broadly useful in diverse fungi for increasing production of a variety of enzymes.
Figures




Similar articles
-
Disruption of Trichoderma reesei cre2, encoding an ubiquitin C-terminal hydrolase, results in increased cellulase activity.BMC Biotechnol. 2011 Nov 9;11:103. doi: 10.1186/1472-6750-11-103. BMC Biotechnol. 2011. PMID: 22070776 Free PMC article.
-
Increased production of biomass-degrading enzymes by double deletion of creA and creB genes involved in carbon catabolite repression in Aspergillus oryzae.J Biosci Bioeng. 2018 Feb;125(2):141-147. doi: 10.1016/j.jbiosc.2017.08.019. Epub 2017 Sep 30. J Biosci Bioeng. 2018. PMID: 28970110
-
Genetic analysis of conidiation regulatory pathways in koji-mold Aspergillus oryzae.Fungal Genet Biol. 2010 Jan;47(1):10-8. doi: 10.1016/j.fgb.2009.10.004. Fungal Genet Biol. 2010. PMID: 19850144
-
Production of toxic metabolites in Aspergillus niger, Aspergillus oryzae, and Trichoderma reesei: justification of mycotoxin testing in food grade enzyme preparations derived from the three fungi.Regul Toxicol Pharmacol. 2004 Apr;39(2):214-28. doi: 10.1016/j.yrtph.2003.09.002. Regul Toxicol Pharmacol. 2004. PMID: 15041150 Review.
-
Impact of Aspergillus oryzae genomics on industrial production of metabolites.Mycopathologia. 2006 Sep;162(3):143-53. doi: 10.1007/s11046-006-0049-2. Mycopathologia. 2006. PMID: 16944282 Review.
Cited by
-
Induction and Repression of Hydrolase Genes in Aspergillus oryzae.Front Microbiol. 2021 May 24;12:677603. doi: 10.3389/fmicb.2021.677603. eCollection 2021. Front Microbiol. 2021. PMID: 34108952 Free PMC article. Review.
-
Improved α-Amylase Production by Dephosphorylation Mutation of CreD, an Arrestin-Like Protein Required for Glucose-Induced Endocytosis of Maltose Permease and Carbon Catabolite Derepression in Aspergillus oryzae.Appl Environ Microbiol. 2017 Jun 16;83(13):e00592-17. doi: 10.1128/AEM.00592-17. Print 2017 Jul 1. Appl Environ Microbiol. 2017. PMID: 28455339 Free PMC article.
-
Regulators of plant biomass degradation in ascomycetous fungi.Biotechnol Biofuels. 2017 Jun 12;10:152. doi: 10.1186/s13068-017-0841-x. eCollection 2017. Biotechnol Biofuels. 2017. PMID: 28616076 Free PMC article. Review.
-
Current Practices for Reference Gene Selection in RT-qPCR of Aspergillus: Outlook and Recommendations for the Future.Genes (Basel). 2021 Jun 24;12(7):960. doi: 10.3390/genes12070960. Genes (Basel). 2021. PMID: 34202507 Free PMC article. Review.
-
Diverse Regulation of the CreA Carbon Catabolite Repressor in Aspergillus nidulans.Genetics. 2016 May;203(1):335-52. doi: 10.1534/genetics.116.187872. Epub 2016 Mar 26. Genetics. 2016. PMID: 27017621 Free PMC article.
References
-
- Ward OP, Qin WM, Dhanjoon J, Ye J, Singh A. 2006. Physiology and biotechnology of Aspergillus. Adv. Appl. Microbiol. 58:1–55 - PubMed
-
- Fleissner A, Dersch P. 2010. Expression and export: recombinant protein production systems for Aspergillus. Appl. Microbiol. Biotechnol. 87:1255–1270 - PubMed
-
- Hynes MJ, Kelly JM. 1977. Pleiotropic mutants of Aspergillus nidulans altered in carbon metabolism. Mol. Gen. Genet. 150:193–204 - PubMed
-
- Kelly JM, Hynes MJ. 1977. Increased and decreased sensitivity to carbon catabolite repression of enzymes of acetate metabolism in mutants of Aspergillus nidulans. Mol. Gen. Genet. 156:87–92 - PubMed
-
- Abdallah BM, Simoes T, Fernandes A, Strauss J, Seiboth B, Sa-Correia I, Kubicek CP. 2000. Glucose does not activate the plasma-membrane-bound H+-ATPase but affects pmaA transcript abundance in Aspergillus nidulans. Arch. Microbiol. 174:340–345 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

