Multiscale kinetic modeling of liposomal Doxorubicin delivery quantifies the role of tumor and drug-specific parameters in local delivery to tumors
- PMID: 23835797
- PMCID: PMC3600732
- DOI: 10.1038/psp.2012.16
Multiscale kinetic modeling of liposomal Doxorubicin delivery quantifies the role of tumor and drug-specific parameters in local delivery to tumors
Abstract
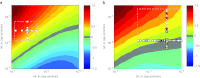
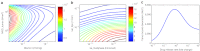
Nanoparticle encapsulation has been used as a means to manipulate the pharmacokinetic (PK) and safety profile of drugs in oncology. Using pegylated liposomal doxorubicin (PLD) vs. conventional doxorubicin as a model system, we developed and experimentally validated a multiscale computational model of liposomal drug delivery. We demonstrated that, for varying tumor transport properties, there is a regimen where liposomal and conventional doxorubicin deliver identical amounts of doxorubicin to tumor cell nuclei. In mice, typical tumor properties consistently favor improved delivery via liposomes relative to free drug. However, in humans, we predict that some tumors will have properties wherein liposomal delivery delivers the identical amount of drug to its target relative to dosing with free drug. The ability to identify tumor types and/or individual patient tumors with high degree of liposome deposition may be critical for optimizing the success of nanoparticle and liposomal anticancer therapeutics.CPT: Pharmacometrics & Systems Pharmacology (2012) 1, e15; doi:10.1038/psp.2012.16; advance online publication 21 November 2012.
Figures
Similar articles
-
Future directions of liposome- and immunoliposome-based cancer therapeutics.Semin Oncol. 2004 Dec;31(6 Suppl 13):196-205. doi: 10.1053/j.seminoncol.2004.08.009. Semin Oncol. 2004. PMID: 15717745 Review.
-
Direct comparison of two pegylated liposomal doxorubicin formulations: is AUC predictive for toxicity and efficacy?J Control Release. 2007 Apr 2;118(2):204-15. doi: 10.1016/j.jconrel.2006.12.002. Epub 2006 Dec 8. J Control Release. 2007. PMID: 17239468
-
Clinical pharmacology of liposomal anthracyclines: focus on pegylated liposomal Doxorubicin.Clin Lymphoma Myeloma. 2008 Feb;8(1):21-32. doi: 10.3816/clm.2008.n.001. Clin Lymphoma Myeloma. 2008. PMID: 18501085 Review.
-
Her2-targeted pegylated liposomal doxorubicin: retention of target-specific binding and cytotoxicity after in vivo passage.J Control Release. 2009 Jun 5;136(2):155-60. doi: 10.1016/j.jconrel.2009.02.002. Epub 2009 Feb 7. J Control Release. 2009. PMID: 19331844
-
Cell-penetrating corosolic acid liposome as a functional carrier for delivering chemotherapeutic drugs.Acta Biomater. 2020 Apr 1;106:301-313. doi: 10.1016/j.actbio.2020.02.013. Epub 2020 Feb 17. Acta Biomater. 2020. PMID: 32081779
Cited by
-
Improving nanotherapy delivery and action through image-guided systems pharmacology.Theranostics. 2020 Jan 1;10(3):968-997. doi: 10.7150/thno.37215. eCollection 2020. Theranostics. 2020. PMID: 31938046 Free PMC article. Review.
-
Delivery of cancer therapeutics to extracellular and intracellular targets: Determinants, barriers, challenges and opportunities.Adv Drug Deliv Rev. 2016 Feb 1;97:280-301. doi: 10.1016/j.addr.2015.12.002. Epub 2015 Dec 11. Adv Drug Deliv Rev. 2016. PMID: 26686425 Free PMC article. Review.
-
Research tools for extrapolating the disposition and pharmacokinetics of nanomaterials from preclinical animals to humans.Theranostics. 2019 May 18;9(11):3365-3387. doi: 10.7150/thno.34509. eCollection 2019. Theranostics. 2019. PMID: 31244958 Free PMC article. Review.
-
Applications of Quantitative Systems Pharmacology in Model-Informed Drug Discovery: Perspective on Impact and Opportunities.CPT Pharmacometrics Syst Pharmacol. 2019 Nov;8(11):777-791. doi: 10.1002/psp4.12463. Epub 2019 Oct 25. CPT Pharmacometrics Syst Pharmacol. 2019. PMID: 31535440 Free PMC article.
-
Physiologically based pharmacokinetic modeling of intravenously administered nanoformulated substances.Drug Deliv Transl Res. 2022 Sep;12(9):2132-2144. doi: 10.1007/s13346-022-01159-w. Epub 2022 May 12. Drug Deliv Transl Res. 2022. PMID: 35551616 Free PMC article.
References
-
- Lefrak E.A., Pitha J., Rosenheim S., &, Gottlieb J.A. A clinicopathologic analysis of adriamycin cardiotoxicity. Cancer. 1973;32:302–314. - PubMed
-
- Colbern G.T., Hiller A.J., Musterer R.S., Pegg E., Henderson I.C., &, Working P.K. Significant increase in antitumor potency of doxorubicin Hc1 by its encapsulation in pegylated liposomes. J. Liposome Res. 1999;9:523.
LinkOut - more resources
Full Text Sources
Miscellaneous

