Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons
- PMID: 23836290
- PMCID: PMC3753484
- DOI: 10.1007/s00401-013-1149-y
Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons
Erratum in
- Acta Neuropathol. 2014 Jun;127(6):941
Abstract
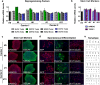
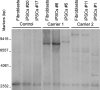
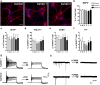
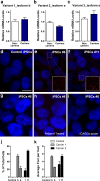
The recently identified GGGGCC repeat expansion in the noncoding region of C9ORF72 is the most common pathogenic mutation in patients with frontotemporal dementia (FTD) or amyotrophic lateral sclerosis (ALS). We generated a human neuronal model and investigated the pathological phenotypes of human neurons containing GGGGCC repeat expansions. Skin biopsies were obtained from two subjects who had >1,000 GGGGCC repeats in C9ORF72 and their respective fibroblasts were used to generate multiple induced pluripotent stem cell (iPSC) lines. After extensive characterization, two iPSC lines from each subject were selected, differentiated into postmitotic neurons, and compared with control neurons to identify disease-relevant phenotypes. Expanded GGGGCC repeats exhibit instability during reprogramming and neuronal differentiation of iPSCs. RNA foci containing GGGGCC repeats were present in some iPSCs, iPSC-derived human neurons and primary fibroblasts. The percentage of cells with foci and the number of foci per cell appeared to be determined not simply by repeat length but also by other factors. These RNA foci do not seem to sequester several major RNA-binding proteins. Moreover, repeat-associated non-ATG (RAN) translation products were detected in human neurons with GGGGCC repeat expansions and these neurons showed significantly elevated p62 levels and increased sensitivity to cellular stress induced by autophagy inhibitors. Our findings demonstrate that key neuropathological features of FTD/ALS with GGGGCC repeat expansions can be recapitulated in iPSC-derived human neurons and also suggest that compromised autophagy function may represent a novel underlying pathogenic mechanism.
Figures


References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- R01 AG038791/AG/NIA NIH HHS/United States
- NS057553/NS/NINDS NIH HHS/United States
- AG026251/AG/NIA NIH HHS/United States
- AG019724/AG/NIA NIH HHS/United States
- NS063964/NS/NINDS NIH HHS/United States
- AG023501/AG/NIA NIH HHS/United States
- P01 AG017216/AG/NIA NIH HHS/United States
- NS077402/NS/NINDS NIH HHS/United States
- P50 AG023501/AG/NIA NIH HHS/United States
- R01 NS063964/NS/NINDS NIH HHS/United States
- P01 AG019724/AG/NIA NIH HHS/United States
- R37 NS057553/NS/NINDS NIH HHS/United States
- R01 NS057553/NS/NINDS NIH HHS/United States
- R01 AG026251/AG/NIA NIH HHS/United States
- R01 NS077402/NS/NINDS NIH HHS/United States
- R56 AG026251/AG/NIA NIH HHS/United States
- AG17216/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

