A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode
- PMID: 23836863
- PMCID: PMC3754605
- DOI: 10.1128/JB.00432-13
A thermoacidophile-specific protein family, DUF3211, functions as a fatty acid carrier with novel binding mode
Abstract
STK_08120 is a member of the thermoacidophile-specific DUF3211 protein family from Sulfolobus tokodaii strain 7. Its molecular function remains obscure, and sequence similarities for obtaining functional remarks are not available. In this study, the crystal structure of STK_08120 was determined at 1.79-Å resolution to predict its probable function using structure similarity searches. The structure adopts an α/β structure of a helix-grip fold, which is found in the START domain proteins with cavities for hydrophobic substrates or ligands. The detailed structural features implied that fatty acids are the primary ligand candidates for STK_08120, and binding assays revealed that the protein bound long-chain saturated fatty acids (>C14) and their trans-unsaturated types with an affinity equal to that for major fatty acid binding proteins in mammals and plants. Moreover, the structure of an STK_08120-myristic acid complex revealed a unique binding mode among fatty acid binding proteins. These results suggest that the thermoacidophile-specific protein family DUF3211 functions as a fatty acid carrier with a novel binding mode.
Figures




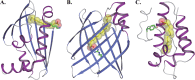
References
-
- Kawarabayasi Y, Hino Y, Horikawa H, Jin-no K, Takahashi M, Sekine M, Baba S, Ankai A, Kosugi H, Hosoyama A, Fukui S, Nagai Y, Nishijima K, Otsuka R, Nakazawa H, Takamiya M, Kato Y, Yoshizawa T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Masuda S, Yanagii M, Nishimura M, Yamagishi A, Oshima T, Kikuchi H. 2001. Complete genome sequence of an aerobic thermoacidophilic crenarchaeon, Sulfolobus tokodaii strain7. DNA Res. 8:123–140 - PubMed
-
- She Q, Singh RK, Confalonieri F, Zivanovic Y, Allard G, Awayez MJ, Chan-Weiher CC, Clausen IG, Curtis BA, De Moors A, Erauso G, Fletcher C, Gordon PM, Heikamp-de Jong I, Jeffries AC, Kozera CJ, Medina N, Peng X, Thi-Ngoc HP, Redder P, Schenk ME, Theriault C, Tolstrup N, Charlebois RL, Doolittle WF, Duguet M, Gaasterland T, Garrett RA, Ragan MA, Sensen CW, Van der Oost J. 2001. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. U. S. A. 98:7835–7840 - PMC - PubMed
-
- Ruepp A, Graml W, Santos-Martinez ML, Koretke KK, Volker C, Mewes HW, Frishman D, Stocker S, Lupas AN, Baumeister W. 2000. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum. Nature 407:508–513 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

