Peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells
- PMID: 23836911
- PMCID: PMC3757184
- DOI: 10.1074/jbc.M113.489674
Peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1)- and estrogen-related receptor (ERR)-induced regulator in muscle 1 (Perm1) is a tissue-specific regulator of oxidative capacity in skeletal muscle cells
Abstract
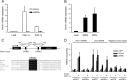
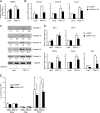
Mitochondrial oxidative metabolism and energy transduction pathways are critical for skeletal and cardiac muscle function. The expression of genes important for mitochondrial biogenesis and oxidative metabolism are under the control of members of the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) family of transcriptional coactivators and the estrogen-related receptor (ERR) subfamily of nuclear receptors. Perturbations in PGC-1 and/or ERR activities have been associated with alterations in capacity for endurance exercise, rates of muscle atrophy, and cardiac function. The mechanism(s) by which PGC-1 and ERR proteins regulate muscle-specific transcriptional programs is not fully understood. We show here that PGC-1α and ERRs induce the expression of a so far uncharacterized muscle-specific protein, PGC-1- and ERR-induced regulator in muscle 1 (Perm1), which regulates the expression of selective PGC-1/ERR target genes. Perm1 is required for the basal as well as PGC-1α-enhanced expression of genes with roles in glucose and lipid metabolism, energy transfer, and contractile function. Silencing of Perm1 in cultured myotubes compromises respiratory capacity and diminishes PGC-1α-induced mitochondrial biogenesis. Our findings support a role for Perm1 acting downstream of PGC-1α and ERRs to regulate muscle-specific pathways important for energy metabolism and contractile function. Elucidating the function of Perm1 may enable novel approaches for the treatment of disorders with compromised skeletal muscle bioenergetics, such as mitochondrial myopathies and age-related/disease-associated muscle atrophies.
Keywords: Bioenergetics; Energy Metabolism; Estrogen-related Receptor; Mitochondrial Biogenesis; Nuclear Receptors; PGC-1α; Skeletal Muscle; Transcription Regulation.
Figures



References
-
- Wallace D. C. (2000) Mitochondrial defects in cardiomyopathy and neuromuscular disease. Am. Heart J. 139, S70–S85 - PubMed
-
- DiMauro S. (2006) Mitochondrial myopathies. Curr. Opin. Rheumatol. 18, 636–641 - PubMed
-
- van Adel B. A., Tarnopolsky M. A. (2009) Metabolic myopathies: update 2009. J. Clin. Neuromuscul. Dis. 10, 97–121 - PubMed
-
- Ingwall J. S. (2009) On the control of metabolic remodeling in mitochondria of the failing heart. Circ. Heart Fail. 2, 275–277 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

