Hidden behind autophagy: the unconventional roles of ATG proteins
- PMID: 23837619
- PMCID: PMC7169877
- DOI: 10.1111/tra.12091
Hidden behind autophagy: the unconventional roles of ATG proteins
Abstract
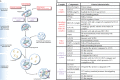
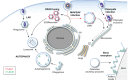
Macroautophagy (hereafter referred to as autophagy) is an evolutionarily conserved intracellular catabolic transport route that generally allows the lysosomal degradation of cytoplasmic components, including bulk cytosol, protein aggregates, damaged or superfluous organelles and invading microbes. Target structures are sequestered by double-membrane vesicles called autophagosomes, which are formed through the concerted action of the autophagy (ATG)-related proteins. Until recently it was assumed that ATG proteins were exclusively involved in autophagy. A growing number of studies, however, have attributed functions to some of them that are distinct from their classical role in autophagosome biogenesis. Autophagy-independent roles of the ATG proteins include the maintenance of cellular homeostasis and resistance to pathogens. For example, they assist and enhance the turnover of dead cells and microbes upon their phagocytic engulfment, and inhibit murine norovirus replication. Moreover, bone resorption by osteoclasts, innate immune regulation triggered by cytoplasmic DNA and the ER-associated degradation regulation all have in common the requirement of a subset of ATG proteins. Microorganisms such as coronaviruses, Chlamydia trachomatis or Brucella abortus have even evolved ways to manipulate autophagy-independent functions of ATG proteins in order to ensure the completion of their intracellular life cycle. Taken together these novel mechanisms add to the repertoire of functions and extend the number of cellular processes involving the ATG proteins.
Keywords: ATG proteins; apoptosis; autophagy; degradation; immunity; infection; pathogens; subversion; unconventional.
© 2013 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

