Multiplexed Illumina sequencing libraries from picogram quantities of DNA
- PMID: 23837789
- PMCID: PMC3711846
- DOI: 10.1186/1471-2164-14-466
Multiplexed Illumina sequencing libraries from picogram quantities of DNA
Abstract
Background: High throughput sequencing is frequently used to discover the location of regulatory interactions on chromatin. However, techniques that enrich DNA where regulatory activity takes place, such as chromatin immunoprecipitation (ChIP), often yield less DNA than optimal for sequencing library preparation. Existing protocols for picogram-scale libraries require concomitant fragmentation of DNA, pre-amplification, or long overnight steps.
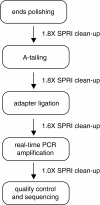
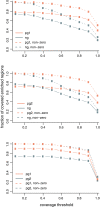
Results: We report a simple and fast library construction method that produces libraries from sub-nanogram quantities of DNA. This protocol yields conventional libraries with barcodes suitable for multiplexed sample analysis on the Illumina platform. We demonstrate the utility of this method by constructing a ChIP-seq library from 100 pg of ChIP DNA that demonstrates equivalent genomic coverage of target regions to a library produced from a larger scale experiment.
Conclusions: Application of this method allows whole genome studies from samples where material or yields are limiting.
Figures

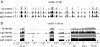
References
-
- Syed F, Grunenwald H, Caruccio N. Next-generation sequencing library preparation: Simultaneous fragmentation and tagging using in vitro transposition. In Nat Methods Application Note. 2009;6 http://www.nature.com/nmeth/journal/v6/n11/full/nmeth.f.272.html.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

