DNA replication origins
- PMID: 23838439
- PMCID: PMC3783049
- DOI: 10.1101/cshperspect.a010116
DNA replication origins
Abstract
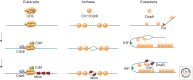
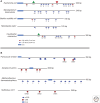
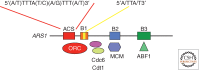
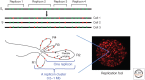
The onset of genomic DNA synthesis requires precise interactions of specialized initiator proteins with DNA at sites where the replication machinery can be loaded. These sites, defined as replication origins, are found at a few unique locations in all of the prokaryotic chromosomes examined so far. However, replication origins are dispersed among tens of thousands of loci in metazoan chromosomes, thereby raising questions regarding the role of specific nucleotide sequences and chromatin environment in origin selection and the mechanisms used by initiators to recognize replication origins. Close examination of bacterial and archaeal replication origins reveals an array of DNA sequence motifs that position individual initiator protein molecules and promote initiator oligomerization on origin DNA. Conversely, the need for specific recognition sequences in eukaryotic replication origins is relaxed. In fact, the primary rule for origin selection appears to be flexibility, a feature that is modulated either by structural elements or by epigenetic mechanisms at least partly linked to the organization of the genome for gene expression.
Figures
References
-
- Aggarwal BD, Calvi BR 2004. Chromatin regulates origin activity in Drosophila follicle cells. Nature 430: 372–376 - PubMed
-
- Beattie TR, Bell SD 2011. Molecular machines in archaeal DNA replication. Curr Opin Chem Biol 15: 614–619 - PubMed
-
- Bell SP, Stillman B 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357: 128–134 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
