DNA replication timing
- PMID: 23838440
- PMCID: PMC3721284
- DOI: 10.1101/cshperspect.a010132
DNA replication timing
Abstract
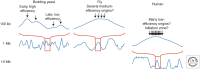
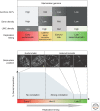
Patterns of replication within eukaryotic genomes correlate with gene expression, chromatin structure, and genome evolution. Recent advances in genome-scale mapping of replication kinetics have allowed these correlations to be explored in many species, cell types, and growth conditions, and these large data sets have allowed quantitative and computational analyses. One striking new correlation to emerge from these analyses is between replication timing and the three-dimensional structure of chromosomes. This correlation, which is significantly stronger than with any single histone modification or chromosome-binding protein, suggests that replication timing is controlled at the level of chromosomal domains. This conclusion dovetails with parallel work on the heterogeneity of origin firing and the competition between origins for limiting activators to suggest a model in which the stochastic probability of individual origin firing is modulated by chromosomal domain structure to produce patterns of replication. Whether these patterns have inherent biological functions or simply reflect higher-order genome structure is an open question.
Figures


References
-
- Agier N, Fischer G 2012. The mutational profile of the yeast genome is shaped by replication. Mol Biol Evol 29: 905–913 - PubMed
-
- Anglana M, Apiou F, Bensimon A, Debatisse M 2003. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell 114: 385–394 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
