Proteolytic characteristics of cathepsin D related to the recognition and cleavage of its target proteins
- PMID: 23840360
- PMCID: PMC3688724
- DOI: 10.1371/journal.pone.0065733
Proteolytic characteristics of cathepsin D related to the recognition and cleavage of its target proteins
Abstract
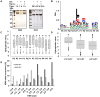
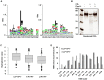
Cathepsin D (CD) plays an important role in both biological and pathological processes, although the cleavage characteristics and substrate selection of CD have yet to be fully explored. We employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) to identify the CD cleavage sites in bovine serum albumin (BSA). We found that the hydrophobic residues at P1 were not only a preferential factor for CD cleavage but that the hydrophobicity at P1' also contributed to CD recognition. The concept of hydrophobic scores of neighbors (HSN) was proposed to describe the hydrophobic microenvironment of CD recognition sites. The survey of CD cleavage characteristics in several proteins suggested that the HSN was a sensitive indicator for judging the favorable sites in peptides for CD cleavage, with HSN values of 0.5-1.0 representing a likely threshold. Ovalbumin (OVA), a protein resistant to CD cleavage in its native state, was easily cleaved by CD after denaturation, and the features of the cleaved peptides were quite similar to those found in BSA, where a higher HSN value indicated greater cleavability. We further conducted two-dimensional gel electrophoresis (2DE) to find more proteins that were insensitive to CD cleavage in CD-knockdown cells. Based on an analysis of secondary and three-dimensional structures, we postulated that intact proteins with a structure consisting of all α-helices would be relatively accessible to CD cleavage.
Conflict of interest statement
Figures


References
-
- Raptis SZ, Shapiro SD, Simmons PM, Cheng AM, Pham CTN (2005) Serine Protease Cathepsin G Regulates Adhesion-Dependent Neutrophil Effector Functions by Modulating Integrin Clustering. Immunity 22: 679–691. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

