Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life
- PMID: 23840569
- PMCID: PMC3695978
- DOI: 10.1371/journal.pone.0066986
Bacterial diversity in meconium of preterm neonates and evolution of their fecal microbiota during the first month of life
Abstract
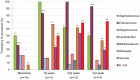
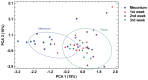
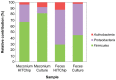
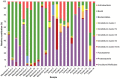
The establishment and succession of bacterial communities in infants may have a profound impact in their health, but information about the composition of meconium microbiota and its evolution in hospitalized preterm infants is scarce. In this context, the objective of this work was to characterize the microbiota of meconium and fecal samples obtained during the first 3 weeks of life from 14 donors using culture and molecular techniques, including DGGE and the Human Intestinal Tract Chip (HITChip) analysis of 16S rRNA amplicons. Culture techniques offer a quantification of cultivable bacteria and allow further study of the isolate, while molecular techniques provide deeper information on bacterial diversity. Culture and HITChip results were very similar but the former showed lower sensitivity. Inter-individual differences were detected in the microbiota profiles although the meconium microbiota was peculiar and distinct from that of fecal samples. Bacilli and other Firmicutes were the main bacteria groups detected in meconium while Proteobacteria dominated in the fecal samples. Culture technique showed that Staphylococcus predominated in meconium and that Enterococcus, together with Gram-negative bacteria such as Escherichia coli, Escherichia fergusonii, Klebsiella pneumoniae and Serratia marcescens, was more abundant in fecal samples. In addition, HITChip results showed the prevalence of bacteria related to Lactobacillus plantarum and Streptococcus mitis in meconium samples whereas those related to Enterococcus, Escherichia coli, Klebsiella pneumoniae and Yersinia predominated in the 3(rd) week feces. This study highlights that spontaneously-released meconium of preterm neonates contains a specific microbiota that differs from that of feces obtained after the first week of life. Our findings indicate that the presence of Serratia was strongly associated with a higher degree of immaturity and other hospital-related parameters, including antibiotherapy and mechanical ventilation.
Conflict of interest statement
Figures



References
-
- Blaut M, Clavel T (2007) Metabolic diversity of the intestinal microbiota: implications for health and disease. J Nutr 137 (3 Suppl 2)751S–755S. - PubMed
-
- Dethlefsen L, Eckburg PB, Bik EM, Relman DA (2006) Assembly of the human intestinal microbiota. Trends Ecol Evol 21: 517–523. - PubMed
-
- Favier CF, de Vos WM, Akkermans AD (2003) Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe 9: 219–229. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

