Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
- PMID: 23840694
- PMCID: PMC3693963
- DOI: 10.1371/journal.pone.0067409
Crystal Structures of the Sec1/Munc18 (SM) Protein Vps33, Alone and Bound to the Homotypic Fusion and Vacuolar Protein Sorting (HOPS) Subunit Vps16*
Abstract
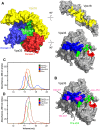
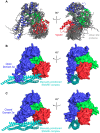
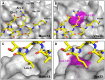
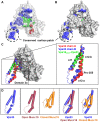
Intracellular membrane fusion requires the regulated assembly of SNARE (soluble N-ethylmaleimide-sensitive factor (NSF) attachment protein receptor) proteins anchored in the apposed membranes. To exert the force required to drive fusion between lipid bilayers, juxtamembrane SNARE motifs zipper into four-helix bundles. Importantly, SNARE function is regulated by additional factors, none more extensively studied than the SM (Sec1/Munc18-like) proteins. SM proteins interact with both individual SNAREs and SNARE complexes, likely chaperoning SNARE complex formation and protecting assembly intermediates from premature disassembly by NSF. Four families of SM proteins have been identified, and representative members of two of these families (Sec1/Munc18 and Sly1) have been structurally characterized. We report here the 2.6 Å resolution crystal structure of an SM protein from the third family, Vps33. Although Vps33 shares with the first two families the same basic three-domain architecture, domain 1 is displaced by 15 Å, accompanied by a 40° rotation. A unique feature of the Vps33 family of SM proteins is that its members function as stable subunits within a multi-subunit tethering complex called HOPS (homotypic fusion and vacuolar protein sorting). Integration into the HOPS complex depends on the interaction between Vps33 and a second HOPS subunit, Vps16. The crystal structure of Vps33 bound to a C-terminal portion of Vps16, also at 2.6 Å resolution, reveals the structural basis for this interaction. Despite the extensive interface between the two HOPS subunits, the conformation of Vps33 is only subtly affected by binding to Vps16.
Conflict of interest statement
Figures


Similar articles
-
Sec1/Munc18 protein Vps33 binds to SNARE domains and the quaternary SNARE complex.Mol Biol Cell. 2012 Dec;23(23):4611-22. doi: 10.1091/mbc.E12-05-0343. Epub 2012 Oct 10. Mol Biol Cell. 2012. PMID: 23051737 Free PMC article.
-
A direct role for the Sec1/Munc18-family protein Vps33 as a template for SNARE assembly.Science. 2015 Sep 4;349(6252):1111-4. doi: 10.1126/science.aac7906. Science. 2015. PMID: 26339030 Free PMC article.
-
The Habc domain of the SNARE Vam3 interacts with the HOPS tethering complex to facilitate vacuole fusion.J Biol Chem. 2015 Feb 27;290(9):5405-13. doi: 10.1074/jbc.M114.631465. Epub 2015 Jan 6. J Biol Chem. 2015. PMID: 25564619 Free PMC article.
-
Tethering the assembly of SNARE complexes.Trends Cell Biol. 2014 Jan;24(1):35-43. doi: 10.1016/j.tcb.2013.09.006. Epub 2013 Oct 9. Trends Cell Biol. 2014. PMID: 24119662 Review.
-
Decoding the interactions of SM proteins with SNAREs.ScientificWorldJournal. 2005 Jun 8;5:471-7. doi: 10.1100/tsw.2005.53. ScientificWorldJournal. 2005. PMID: 15962202 Free PMC article. Review.
Cited by
-
CAPS and Munc13: CATCHRs that SNARE Vesicles.Front Endocrinol (Lausanne). 2013 Dec 4;4:187. doi: 10.3389/fendo.2013.00187. Front Endocrinol (Lausanne). 2013. PMID: 24363652 Free PMC article. Review.
-
Munc18 and Munc13 serve as a functional template to orchestrate neuronal SNARE complex assembly.Nat Commun. 2019 Jan 8;10(1):69. doi: 10.1038/s41467-018-08028-6. Nat Commun. 2019. PMID: 30622273 Free PMC article.
-
Recruitment of VPS33A to HOPS by VPS16 Is Required for Lysosome Fusion with Endosomes and Autophagosomes.Traffic. 2015 Jul;16(7):727-42. doi: 10.1111/tra.12283. Epub 2015 Apr 30. Traffic. 2015. PMID: 25783203 Free PMC article.
-
Deep brain stimulation of the subthalamic nucleus preferentially alters the translational profile of striatopallidal neurons in an animal model of Parkinson's disease.Front Cell Neurosci. 2015 Jun 9;9:221. doi: 10.3389/fncel.2015.00221. eCollection 2015. Front Cell Neurosci. 2015. PMID: 26106299 Free PMC article.
-
Chaperoning SNARE assembly and disassembly.Nat Rev Mol Cell Biol. 2016 Aug;17(8):465-79. doi: 10.1038/nrm.2016.65. Epub 2016 Jun 15. Nat Rev Mol Cell Biol. 2016. PMID: 27301672 Free PMC article. Review.
References
-
- Rizo J, Sudhof TC (2012) The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices–guilty as charged? Ann Rev Cell Dev Biol 28: 279–308. - PubMed
-
- Yu I, Hughson FM (2010) Tethering factors as organizers of intracellular vesicular traffic. Ann Rev Cell Dev Biol 26: 137–156. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

