Periodic distribution of a putative nucleosome positioning motif in human, nonhuman primates, and archaea: mutual information analysis
- PMID: 23841049
- PMCID: PMC3691935
- DOI: 10.1155/2013/963956
Periodic distribution of a putative nucleosome positioning motif in human, nonhuman primates, and archaea: mutual information analysis
Abstract
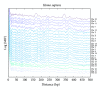
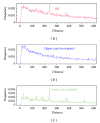
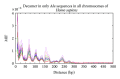
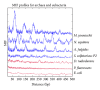
Recently, Trifonov's group proposed a 10-mer DNA motif YYYYYRRRRR as a solution of the long-standing problem of sequence-based nucleosome positioning. To test whether this generic decamer represents a biological meaningful signal, we compare the distribution of this motif in primates and Archaea, which are known to contain nucleosomes, and in Eubacteria, which do not possess nucleosomes. The distribution of the motif is analyzed by the mutual information function (MIF) with a shifted version of itself (MIF profile). We found common features in the patterns of this generic decamer on MIF profiles among primate species, and interestingly we found conspicuous but dissimilar MIF profiles for each Archaea tested. The overall MIF profiles for each chromosome in each primate species also follow a similar pattern. Trifonov's generic decamer may be a highly conserved motif for the nucleosome positioning, but we argue that this is not the only motif. The distribution of this generic decamer exhibits previously unidentified periodicities, which are associated to highly repetitive sequences in the genome. Alu repetitive elements contribute to the most fundamental structure of nucleosome positioning in higher Eukaryotes. In some regions of primate chromosomes, the distribution of the decamer shows symmetrical patterns including inverted repeats.
Figures








Similar articles
-
Effects of Alu elements on global nucleosome positioning in the human genome.BMC Genomics. 2010 May 17;11:309. doi: 10.1186/1471-2164-11-309. BMC Genomics. 2010. PMID: 20478020 Free PMC article.
-
Nucleosome positioning by human Alu elements in chromatin.J Biol Chem. 1995 Apr 28;270(17):10091-6. doi: 10.1074/jbc.270.17.10091. J Biol Chem. 1995. PMID: 7730313
-
Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs.BMC Genomics. 2013 Jun 10;14:391. doi: 10.1186/1471-2164-14-391. BMC Genomics. 2013. PMID: 23758892 Free PMC article.
-
DNA repeats and archaeal nucleosome positioning.Res Microbiol. 1999 Nov-Dec;150(9-10):701-9. doi: 10.1016/s0923-2508(99)00122-9. Res Microbiol. 1999. PMID: 10673008 Review.
-
Active nucleosome positioning beyond intrinsic biophysics is revealed by in vitro reconstitution.Biochem Soc Trans. 2012 Apr;40(2):377-82. doi: 10.1042/BST20110730. Biochem Soc Trans. 2012. PMID: 22435815 Review.
Cited by
-
Database of Periodic DNA Regions in Major Genomes.Biomed Res Int. 2017;2017:7949287. doi: 10.1155/2017/7949287. Epub 2017 Jan 15. Biomed Res Int. 2017. PMID: 28182099 Free PMC article.
-
A Proposal for the RNAome at the Dawn of the Last Universal Common Ancestor.Genes (Basel). 2024 Sep 11;15(9):1195. doi: 10.3390/genes15091195. Genes (Basel). 2024. PMID: 39336786 Free PMC article.
References
-
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421(6921):448–453. - PubMed
-
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. Journal of Molecular Biology. 1997;272(3):301–311. - PubMed
-
- Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. Journal of Molecular Biology. 2002;319(5):1097–1113. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

