Structure and dynamics of urea/water mixtures investigated by vibrational spectroscopy and molecular dynamics simulation
- PMID: 23841646
- PMCID: PMC3808478
- DOI: 10.1021/jp4037217
Structure and dynamics of urea/water mixtures investigated by vibrational spectroscopy and molecular dynamics simulation
Abstract
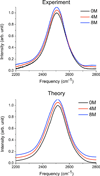
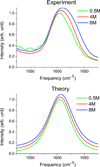
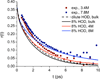
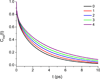
Urea/water is an archetypical "biological" mixture and is especially well-known for its relevance to protein thermodynamics as urea acts as a protein denaturant at high concentration. This behavior has given rise to an extended debate concerning urea's influence on water structure. On the basis of a variety of methods and of definitions of the water structure, urea has been variously described as a structure-breaker, a structure-maker, or as remarkably neutral toward water. Because of its sensitivity to microscopic structure and dynamics, vibrational spectroscopy can help resolve these debates. We report experimental and theoretical spectroscopic results for the OD stretch of HOD/H2O/urea mixtures (linear IR, 2DIR, and pump-probe anisotropy decay) and for the CO stretch of urea-D4/D2O mixtures (linear IR only). Theoretical results are obtained using existing approaches for water and a modification of a frequency map developed for acetamide. All absorption spectra are remarkably insensitive to urea concentration, consistent with the idea that urea only very weakly perturbs the water structure. Both this work and experiments by Rezus and Bakker, however, show that water's rotational dynamics are slowed down by urea. Analysis of the simulations casts doubt on the suggestion that urea immobilizes particular doubly hydrogen bonded water molecules.
Conflict of interest statement
The authors declare no competing financial interest.
Figures

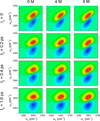
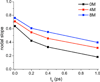
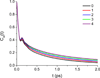
Similar articles
-
Molecular dynamics simulation of aqueous urea solution: is urea a structure breaker?J Phys Chem B. 2014 Oct 9;118(40):11757-68. doi: 10.1021/jp505147u. Epub 2014 Sep 26. J Phys Chem B. 2014. PMID: 25257762
-
Computational Vibrational Spectroscopy of HDO in Osmolyte-Water Solutions.J Phys Chem A. 2016 Jul 28;120(29):5874-86. doi: 10.1021/acs.jpca.6b06305. Epub 2016 Jul 18. J Phys Chem A. 2016. PMID: 27341918
-
A single methyl group drastically changes urea's hydration dynamics.J Chem Phys. 2022 Apr 28;156(16):164504. doi: 10.1063/5.0085461. J Chem Phys. 2022. PMID: 35490020
-
Two-dimensional Infrared Spectroscopy Reveals Better Insights of Structure and Dynamics of Protein.Molecules. 2021 Nov 16;26(22):6893. doi: 10.3390/molecules26226893. Molecules. 2021. PMID: 34833985 Free PMC article. Review.
-
Two-dimensional infrared spectroscopy of intermolecular hydrogen bonds in the condensed phase.Acc Chem Res. 2009 Sep 15;42(9):1220-8. doi: 10.1021/ar900006u. Acc Chem Res. 2009. PMID: 19425543 Review.
Cited by
-
Proteins in binary solvents.Biophys Rev. 2016 Jun;8(2):87-106. doi: 10.1007/s12551-016-0193-y. Epub 2016 Mar 18. Biophys Rev. 2016. PMID: 28510051 Free PMC article. Review.
-
Water-Induced Restructuring of the Surface of a Deep Eutectic Solvent.J Phys Chem Lett. 2022 Jan 20;13(2):634-641. doi: 10.1021/acs.jpclett.1c03907. Epub 2022 Jan 12. J Phys Chem Lett. 2022. PMID: 35020401 Free PMC article.
-
Dispersion interactions between urea and nucleobases contribute to the destabilization of RNA by urea in aqueous solution.J Phys Chem B. 2015 Mar 5;119(9):3755-61. doi: 10.1021/jp512414f. Epub 2015 Feb 23. J Phys Chem B. 2015. PMID: 25668757 Free PMC article.
-
Remdesivir Strongly Binds to RNA-Dependent RNA Polymerase, Membrane Protein, and Main Protease of SARS-CoV-2: Indication From Molecular Modeling and Simulations.Front Pharmacol. 2021 Jul 7;12:710778. doi: 10.3389/fphar.2021.710778. eCollection 2021. Front Pharmacol. 2021. PMID: 34305617 Free PMC article.
-
Time-dependent X-ray diffraction studies on urea/hen egg white lysozyme complexes reveal structural changes that indicate onset of denaturation.Sci Rep. 2016 Aug 30;6:32277. doi: 10.1038/srep32277. Sci Rep. 2016. PMID: 27573790 Free PMC article.
References
-
- Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. Living with Water Stress: Evolution of Osmolyte Systems. Science. 1982;217:1214–1222. - PubMed
-
- Vanzi F, Madan B, Sharp K. Effect of the Protein Denaturants Urea and Guanidinium on Water Structure: A Structural and Thermodynamic Study. J. Am. Chem. Soc. 1998;120:10748–10753.
-
- Kallies B. Coupling of Solvent and Solute Dynamics—Molecular Dynamics Simulations of Aqueous Urea Solutions with Different Intramolecular Potentials. Phys. Chem. Chem. Phys. 2002;4:86–95.
-
- Mountain RD, Thirumalai D. Importance of Excluded Volume on the Solvation of Urea in Water. J. Phys. Chem. B. 2004;108:6826–6831.
-
- Felitsky DJ, Record MT. Application of the Local-Bulk Partitioning and Competitive Binding Models to Interpret Preferential Interactions of Glycine Betaine and Urea with Protein Surface. Biochemistry. 2004;43:9276–9288. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

