RrmA regulates the stability of specific transcripts in response to both nitrogen source and oxidative stress
- PMID: 23841692
- PMCID: PMC4282371
- DOI: 10.1111/mmi.12324
RrmA regulates the stability of specific transcripts in response to both nitrogen source and oxidative stress
Abstract
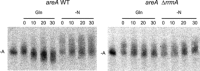
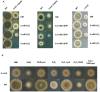
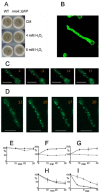
Differential regulation of transcript stability is an effective means by which an organism can modulate gene expression. A well-characterized example is glutamine signalled degradation of specific transcripts in Aspergillus nidulans. In the case of areA, which encodes a wide-domain transcription factor mediating nitrogen metabolite repression, the signal is mediated through a highly conserved region of the 3' UTR. Utilizing this RNA sequence we isolated RrmA, an RNA recognition motif protein. Disruption of the respective gene led to loss of both glutamine signalled transcript degradation as well as nitrate signalled stabilization of niaD mRNA. However, nitrogen starvation was shown to act independently of RrmA in stabilizing certain transcripts. RrmA was also implicated in the regulation of arginine catabolism gene expression and the oxidative stress responses at the level of mRNA stability. ΔrrmA mutants are hypersensitive to oxidative stress. This phenotype correlates with destabilization of eifE and dhsA mRNA. eifE encodes eIF5A, a translation factor within which a conserved lysine is post-translationally modified to hypusine, a process requiring DhsA. Intriguingly, for specific transcripts RrmA mediates both stabilization and destabilization and the specificity of the signals transduced is transcript dependent, suggesting it acts in consort with other factors which differ between transcripts.
© 2013 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures



Similar articles
-
Arginine catabolism in Aspergillus nidulans is regulated by the rrmA gene coding for the RNA-binding protein.Fungal Genet Biol. 2007 Dec;44(12):1285-97. doi: 10.1016/j.fgb.2007.07.001. Epub 2007 Jul 20. Fungal Genet Biol. 2007. PMID: 17719249
-
Opposing signals differentially regulate transcript stability in Aspergillus nidulans.Mol Microbiol. 2006 Oct;62(2):509-19. doi: 10.1111/j.1365-2958.2006.05383.x. Mol Microbiol. 2006. PMID: 17020584
-
Characterization of nitrogen metabolite signalling in Aspergillus via the regulated degradation of areA mRNA.Mol Microbiol. 2001 Oct;42(1):269-77. doi: 10.1046/j.1365-2958.2001.02636.x. Mol Microbiol. 2001. PMID: 11679084
-
Nitrogen regulation in fungi.Antonie Van Leeuwenhoek. 1994;65(3):169-77. doi: 10.1007/BF00871943. Antonie Van Leeuwenhoek. 1994. PMID: 7847882 Review.
-
Cytoplasmic mRNA 3' tagging in eukaryotes: does it spell the end?Biochem Soc Trans. 2012 Aug;40(4):810-4. doi: 10.1042/BST20120068. Biochem Soc Trans. 2012. PMID: 22817739 Review.
Cited by
-
Regulation of nutrient utilization in filamentous fungi.Appl Microbiol Biotechnol. 2023 Oct;107(19):5873-5898. doi: 10.1007/s00253-023-12680-4. Epub 2023 Aug 4. Appl Microbiol Biotechnol. 2023. PMID: 37540250 Free PMC article. Review.
-
Functions of PUF Family RNA-Binding Proteins in Aspergillus nidulans.J Microbiol Biotechnol. 2021 May 28;31(5):676-685. doi: 10.4014/jmb.2101.01011. J Microbiol Biotechnol. 2021. PMID: 33746193 Free PMC article.
-
Expression of a Structural Protein of the Mycovirus FgV-ch9 Negatively Affects the Transcript Level of a Novel Symptom Alleviation Factor and Causes Virus Infection-Like Symptoms in Fusarium graminearum.J Virol. 2018 Aug 16;92(17):e00326-18. doi: 10.1128/JVI.00326-18. Print 2018 Sep 1. J Virol. 2018. PMID: 29899100 Free PMC article.
-
The role of the GATA transcription factor AreB in regulation of nitrogen and carbon metabolism in Aspergillus nidulans.FEMS Microbiol Lett. 2019 Mar 1;366(6):fnz066. doi: 10.1093/femsle/fnz066. FEMS Microbiol Lett. 2019. PMID: 30939206 Free PMC article.
References
-
- Asano Y, Hagiwara D, Yamashino T. Mizuno T. Characterization of the bZip-type transcription factor NapA with reference to oxidative stress response in Aspergillus nidulans. Biosci Biotechnol Biochem. 2007;71:1800–1803. - PubMed
-
- Baker CS, Morozov I, Suzuki K, Romeo T. Babitzke P. CsrA regulates glycogen biosynthesis by preventing translation of glgC in Escherichia coli. Mol Microbiol. 2002;44:1599–1610. - PubMed
-
- Beelman CA. Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J Biol Chem. 1994;269:9687–9692. - PubMed
-
- Bevilacqua A, Ceriani MC, Capaccioli S. Nicolin A. Post-transcriptional regulation of gene expression by degradation of messenger RNAs. J Cell Physiol. 2003;195:356–372. - PubMed
-
- Borsuk P, Przykorska A, Blachnio K, Koper M, Pawlowicz JM, Pekala M. Weglenski P. L-arginine influences the structure and function of arginase mRNA in Aspergillus nidulans. Biol Chem. 2007;388:135–144. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

