Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development
- PMID: 23842086
- PMCID: PMC3725088
- DOI: 10.1073/pnas.1221104110
Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development
Erratum in
- Proc Natl Acad Sci U S A. 2014 Feb 25;111(8):3195
Abstract
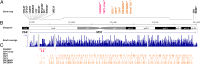
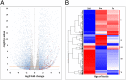
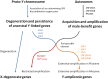
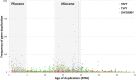
The male-specific region of the mammalian Y chromosome (MSY) contains clusters of genes essential for male reproduction. The highly repetitive and degenerative nature of the Y chromosome impedes genomic and transcriptomic characterization. Although the Y chromosome sequence is available for the human, chimpanzee, and macaque, little is known about the annotation and transcriptome of nonprimate MSY. Here, we investigated the transcriptome of the MSY in cattle by direct testis cDNA selection and RNA-seq approaches. The bovine MSY differs radically from the primate Y chromosomes with respect to its structure, gene content, and density. Among the 28 protein-coding genes/families identified on the bovine MSY (12 single- and 16 multicopy genes), 16 are bovid specific. The 1,274 genes identified in this study made the bovine MSY gene density the highest in the genome; in comparison, primate MSYs have only 31-78 genes. Our results, along with the highly transcriptional activities observed from these Y-chromosome genes and 375 additional noncoding RNAs, challenge the widely accepted hypothesis that the MSY is gene poor and transcriptionally inert. The bovine MSY genes are predominantly expressed and are differentially regulated during the testicular development. Synonymous substitution rate analyses of the multicopy MSY genes indicated that two major periods of expansion occurred during the Miocene and Pliocene, contributing to the adaptive radiation of bovids. The massive amplification and vigorous transcription suggest that the MSY serves as a genomic niche regulating male reproduction during bovid expansion.
Keywords: evolution; expression; gene amplification; transposition.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
No bull: upholding community standards in public sharing of biological datasets.Proc Natl Acad Sci U S A. 2013 Nov 12;110(46):E4277. doi: 10.1073/pnas.1315122110. Epub 2013 Oct 30. Proc Natl Acad Sci U S A. 2013. PMID: 24173037 Free PMC article. No abstract available.
-
Reply to Hughes et al.: No bullying: Publications comply with international standards.Proc Natl Acad Sci U S A. 2013 Nov 12;110(46):E4278. doi: 10.1073/pnas.1316664110. Proc Natl Acad Sci U S A. 2013. PMID: 24367808 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

