Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression
- PMID: 23843461
- PMCID: PMC3757183
- DOI: 10.1074/jbc.M113.484733
Lacto-N-biosidase encoded by a novel gene of Bifidobacterium longum subspecies longum shows unique substrate specificity and requires a designated chaperone for its active expression
Abstract
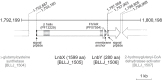
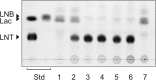
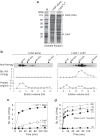
Infant gut-associated bifidobacteria possess species-specific enzymatic sets to assimilate human milk oligosaccharides, and lacto-N-biosidase (LNBase) is a key enzyme that degrades lacto-N-tetraose (Galβ1-3GlcNAcβ1-3Galβ1-4Glc), the main component of human milk oligosaccharides, to lacto-N-biose I (Galβ1-3GlcNAc) and lactose. We have previously identified LNBase activity in Bifidobacterium bifidum and some strains of Bifidobacterium longum subsp. longum (B. longum). Subsequently, we isolated a glycoside hydrolase family 20 (GH20) LNBase from B. bifidum; however, the genome of the LNBase(+) strain of B. longum contains no GH20 LNBase homolog. Here, we reveal that locus tags BLLJ_1505 and BLLJ_1506 constitute LNBase from B. longum JCM1217. The gene products, designated LnbX and LnbY, respectively, showed no sequence similarity to previously characterized proteins. The purified enzyme, which consisted of LnbX only, hydrolyzed via a retaining mechanism the GlcNAcβ1-3Gal linkage in lacto-N-tetraose, lacto-N-fucopentaose I (Fucα1-2Galβ1-3GlcNAcβ1-3Galβ1-4Glc), and sialyllacto-N-tetraose a (Neu5Acα2-3Galβ1-3GlcNAcβ1-3Galβ1-4Gal); the latter two are not hydrolyzed by GH20 LNBase. Among the chromogenic substrates examined, the enzyme acted on p-nitrophenyl (pNP)-β-lacto-N-bioside I (Galβ1-3GlcNAcβ-pNP) and GalNAcβ1-3GlcNAcβ-pNP. GalNAcβ1-3GlcNAcβ linkage has been found in O-mannosyl glycans of α-dystroglycan. Therefore, the enzyme may serve as a new tool for examining glycan structures. In vitro refolding experiments revealed that LnbY and metal ions (Ca(2+) and Mg(2+)) are required for proper folding of LnbX. The LnbX and LnbY homologs have been found only in B. bifidum, B. longum, and a few gut microbes, suggesting that the proteins have evolved in specialized niches.
Keywords: Bacterial Metabolism; Carbohydrate Metabolism; Glycobiology; Glycoside Hydrolases; Microbiology; Oligosaccharide.
Figures




Similar articles
-
Bifidobacterium bifidum lacto-N-biosidase, a critical enzyme for the degradation of human milk oligosaccharides with a type 1 structure.Appl Environ Microbiol. 2008 Jul;74(13):3996-4004. doi: 10.1128/AEM.00149-08. Epub 2008 May 9. Appl Environ Microbiol. 2008. PMID: 18469123 Free PMC article.
-
Enzymatic lacto-N-biose elongation of human milk oligosaccharides with the GH136 lacto-N-biosidase LnbX engineered for improved transglycosylation.Enzyme Microb Technol. 2025 Sep;189:110660. doi: 10.1016/j.enzmictec.2025.110660. Epub 2025 May 4. Enzyme Microb Technol. 2025. PMID: 40328212
-
Molecular Insight into Evolution of Symbiosis between Breast-Fed Infants and a Member of the Human Gut Microbiome Bifidobacterium longum.Cell Chem Biol. 2017 Apr 20;24(4):515-524.e5. doi: 10.1016/j.chembiol.2017.03.012. Epub 2017 Apr 6. Cell Chem Biol. 2017. PMID: 28392148
-
Structure and evolution of the bifidobacterial carbohydrate metabolism proteins and enzymes.Biochem Soc Trans. 2021 Apr 30;49(2):563-578. doi: 10.1042/BST20200163. Biochem Soc Trans. 2021. PMID: 33666221 Free PMC article. Review.
-
Bifidobacterial enzymes involved in the metabolism of human milk oligosaccharides.Adv Nutr. 2012 May 1;3(3):422S-9S. doi: 10.3945/an.111.001420. Adv Nutr. 2012. PMID: 22585921 Free PMC article. Review.
Cited by
-
Comparative Genomics Revealed Genetic Diversity and Species/Strain-Level Differences in Carbohydrate Metabolism of Three Probiotic Bifidobacterial Species.Int J Genomics. 2015;2015:567809. doi: 10.1155/2015/567809. Epub 2015 Jul 5. Int J Genomics. 2015. PMID: 26236711 Free PMC article.
-
Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum.Sci Rep. 2018 Sep 18;8(1):13958. doi: 10.1038/s41598-018-32080-3. Sci Rep. 2018. PMID: 30228375 Free PMC article.
-
Bifidobacterium breve UCC2003 metabolises the human milk oligosaccharides lacto-N-tetraose and lacto-N-neo-tetraose through overlapping, yet distinct pathways.Sci Rep. 2016 Dec 8;6:38560. doi: 10.1038/srep38560. Sci Rep. 2016. PMID: 27929046 Free PMC article.
-
Determining the metabolic fate of human milk oligosaccharides: it may just be more complex than you think?Gut Microbiome (Camb). 2022 Sep 7;3:e9. doi: 10.1017/gmb.2022.8. eCollection 2022. Gut Microbiome (Camb). 2022. PMID: 39295778 Free PMC article. Review.
-
Variation in the Conservation of Species-Specific Gene Sets for HMO Degradation and Its Effects on HMO Utilization in Bifidobacteria.Nutrients. 2024 Jun 15;16(12):1893. doi: 10.3390/nu16121893. Nutrients. 2024. PMID: 38931248 Free PMC article.
References
-
- Mitsuoka T. (1988) Intestinal flora and host. Asian Med. J. 31, 400–409
-
- Penders J., Thijs C., Vink C., Stelma F. F., Snijders B., Kummeling I., van den Brandt P. A., Stobberingh E. E. (2006) Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521 - PubMed
-
- Kunz C., Rudloff S., Baier W., Klein N., Strobel S. (2000) Oligosaccharides in human milk. Structural, functional, and metabolic aspects. Annu. Rev. Nutr. 20, 699–722 - PubMed
-
- Bode L. (2006) Recent advances on structure, metabolism, and function of human milk oligosaccharides. J. Nutr. 136, 2127–2130 - PubMed
-
- Newburg D. S., Neubauer S. H. (1995) Carbohydrates in milk. in Handbook of Milk Composition (Jensen R. G., ed) pp. 273–349, Academic Press, Inc., New York
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

