14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes
- PMID: 23843611
- PMCID: PMC3772389
- DOI: 10.1242/jcs.126102
14-3-3 protein targets misfolded chaperone-associated proteins to aggresomes
Abstract
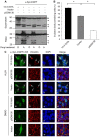
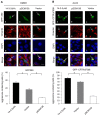
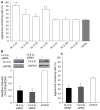
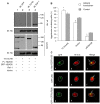
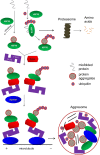
The aggresome is a key cytoplasmic organelle for sequestration and clearance of toxic protein aggregates. Although loading misfolded proteins cargos to dynein motors has been recognized as an important step in the aggresome formation process, the molecular machinery that mediates the association of cargos with the dynein motor is poorly understood. Here, we report a new aggresome-targeting pathway that involves isoforms of 14-3-3, a family of conserved regulatory proteins. 14-3-3 interacts with both the dynein-intermediate chain (DIC) and an Hsp70 co-chaperone Bcl-2-associated athanogene 3 (BAG3), thereby recruiting chaperone-associated protein cargos to dynein motors for their transport to aggresomes. This molecular cascade entails functional dimerization of 14-3-3, which we show to be crucial for the formation of aggresomes in both yeast and mammalian cells. These results suggest that 14-3-3 functions as a molecular adaptor to promote aggresomal targeting of misfolded protein aggregates and may link such complexes to inclusion bodies observed in various neurodegenerative diseases.
Keywords: 14-3-3; Adaptor; Aggresome; BAG3; Dynein.
Figures



Similar articles
-
14-3-3 and aggresome formation: implications in neurodegenerative diseases.Prion. 2014 Mar-Apr;8(2):173-7. doi: 10.4161/pri.28123. Epub 2014 Feb 18. Prion. 2014. PMID: 24549097 Free PMC article. Review.
-
BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins.EMBO Rep. 2011 Feb;12(2):149-56. doi: 10.1038/embor.2010.203. Epub 2011 Jan 21. EMBO Rep. 2011. PMID: 21252941 Free PMC article.
-
BAG3 and friends: co-chaperones in selective autophagy during aging and disease.Autophagy. 2011 Jul;7(7):795-8. doi: 10.4161/auto.7.7.15844. Autophagy. 2011. PMID: 21681022
-
Cytoplasmic dynein/dynactin mediates the assembly of aggresomes.Cell Motil Cytoskeleton. 2002 Sep;53(1):26-38. doi: 10.1002/cm.10057. Cell Motil Cytoskeleton. 2002. PMID: 12211113
-
Aggresome formation and neurodegenerative diseases: therapeutic implications.Curr Med Chem. 2008;15(1):47-60. doi: 10.2174/092986708783330692. Curr Med Chem. 2008. PMID: 18220762 Free PMC article. Review.
Cited by
-
Deciphering the Structure and Formation of Amyloids in Neurodegenerative Diseases With Chemical Biology Tools.Front Chem. 2022 May 12;10:886382. doi: 10.3389/fchem.2022.886382. eCollection 2022. Front Chem. 2022. PMID: 35646824 Free PMC article. Review.
-
Interaction of SQSTM1 with the motor protein dynein--SQSTM1 is required for normal dynein function and trafficking.J Cell Sci. 2014 Sep 15;127(Pt 18):4052-63. doi: 10.1242/jcs.152363. Epub 2014 Jul 11. J Cell Sci. 2014. PMID: 25015291 Free PMC article.
-
BIS targeting induces cellular senescence through the regulation of 14-3-3 zeta/STAT3/SKP2/p27 in glioblastoma cells.Cell Death Dis. 2014 Nov 20;5(11):e1537. doi: 10.1038/cddis.2014.501. Cell Death Dis. 2014. PMID: 25412315 Free PMC article.
-
MARK2 phosphorylates KIF13A at a 14-3-3 binding site to polarize vesicular transport of transferrin receptor within dendrites.Proc Natl Acad Sci U S A. 2024 May 14;121(20):e2316266121. doi: 10.1073/pnas.2316266121. Epub 2024 May 6. Proc Natl Acad Sci U S A. 2024. PMID: 38709923 Free PMC article.
-
The Role of HSPB8, a Component of the Chaperone-Assisted Selective Autophagy Machinery, in Cancer.Cells. 2021 Feb 5;10(2):335. doi: 10.3390/cells10020335. Cells. 2021. PMID: 33562660 Free PMC article. Review.
References
-
- Chen H. K., Fernandez-Funez P., Acevedo S. F., Lam Y. C., Kaytor M. D., Fernandez M. H., Aitken A., Skoulakis E. M., Orr H. T., Botas J. et al.(2003). Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell 113, 457–468 10.1016/S0092-8674(03)00349-0 - DOI - PubMed
-
- Datta S. R., Katsov A., Hu L., Petros A., Fesik S. W., Yaffe M. B., Greenberg M. E. (2000). 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol. Cell 6, 41–51 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

