The ire-1 ER stress-response pathway is required for normal secretory-protein metabolism in C. elegans
- PMID: 23843615
- PMCID: PMC3772388
- DOI: 10.1242/jcs.123000
The ire-1 ER stress-response pathway is required for normal secretory-protein metabolism in C. elegans
Abstract
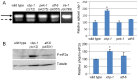
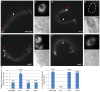
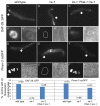
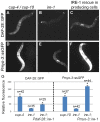
The unfolded protein response (UPR) allows cells to cope with endoplasmic reticulum (ER) stress by adjusting the capacity of the ER to the load of ER-associated tasks. The UPR is important for maintaining ER homeostasis under extreme ER stress. UPR genes are important under normal growth conditions as well, but what they are required for under these conditions is less clear. Using C. elegans, we show that the ire-1/xbp-1 arm of the UPR plays a crucial role in maintaining ER plasticity and function also in the absence of external ER stress. We find that during unstressed growth conditions, loss of ire-1 or xbp-1 compromises basic ER functions required for the metabolism of secreted proteins, including translation, folding and secretion. Notably, by compromising ER-associated degradation (ERAD) and phagocytosis, loss of ire-1 hinders the clearance of misfolded proteins from the ER as well as the clearance of proteins that were secreted into the pseudocoleom. Whereas the basal activity of the UPR is beneficial under normal conditions, it accelerates the pathology caused by toxic Aβ protein in a C. elegans model of Alzheimer's disease. Taken together, our findings indicate that UPR genes are critical for maintaining secretory protein metabolism under normal growth conditions.
Keywords: Alzheimer's disease; Caenorhabditis elegans; Coelomocytes; ER stress; ERAD; UPR.
Figures
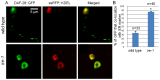
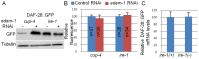
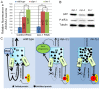
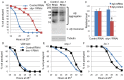
Similar articles
-
Endoplasmic Reticulum Homeostasis Is Modulated by the Forkhead Transcription Factor FKH-9 During Infection of Caenorhabditis elegans.Genetics. 2018 Dec;210(4):1329-1337. doi: 10.1534/genetics.118.301450. Epub 2018 Oct 4. Genetics. 2018. PMID: 30287474 Free PMC article.
-
RNA surveillance is required for endoplasmic reticulum homeostasis.Proc Natl Acad Sci U S A. 2012 May 22;109(21):8079-84. doi: 10.1073/pnas.1110589109. Epub 2012 May 4. Proc Natl Acad Sci U S A. 2012. PMID: 22562797 Free PMC article.
-
Physiological IRE-1-XBP-1 and PEK-1 signaling in Caenorhabditis elegans larval development and immunity.PLoS Genet. 2011 Nov;7(11):e1002391. doi: 10.1371/journal.pgen.1002391. Epub 2011 Nov 17. PLoS Genet. 2011. PMID: 22125500 Free PMC article.
-
Integrated signaling system under endoplasmic reticulum stress in eukaryotic microorganisms.Appl Microbiol Biotechnol. 2021 Jun;105(12):4805-4818. doi: 10.1007/s00253-021-11380-1. Epub 2021 Jun 9. Appl Microbiol Biotechnol. 2021. PMID: 34106312 Review.
-
Endoplasmic reticulum stress, degeneration of pancreatic islet β-cells, and therapeutic modulation of the unfolded protein response in diabetes.Mol Metab. 2019 Sep;27S(Suppl):S60-S68. doi: 10.1016/j.molmet.2019.06.012. Mol Metab. 2019. PMID: 31500832 Free PMC article. Review.
Cited by
-
Restoration of Proteostasis in the Endoplasmic Reticulum Reverses an Inflammation-Like Response to Cytoplasmic DNA in Caenorhabditis elegans.Genetics. 2019 Aug;212(4):1259-1278. doi: 10.1534/genetics.119.302422. Epub 2019 Jun 27. Genetics. 2019. PMID: 31248887 Free PMC article.
-
Expression of hepatitis B virus surface antigens induces defective gonad phenotypes in Caenorhabditis elegans.World J Virol. 2017 Feb 12;6(1):17-25. doi: 10.5501/wjv.v6.i1.17. World J Virol. 2017. PMID: 28239568 Free PMC article.
-
ER stress and the unfolded protein response in neurodegeneration.Nat Rev Neurol. 2017 Aug;13(8):477-491. doi: 10.1038/nrneurol.2017.99. Epub 2017 Jul 21. Nat Rev Neurol. 2017. PMID: 28731040 Review.
-
ER-stress in Alzheimer's disease: turning the scale?Am J Neurodegener Dis. 2013 Nov 29;2(4):247-65. Am J Neurodegener Dis. 2013. PMID: 24319643 Free PMC article. Review.
-
Simple model systems reveal conserved mechanisms of Alzheimer's disease and related tauopathies.Mol Neurodegener. 2023 Nov 10;18(1):82. doi: 10.1186/s13024-023-00664-x. Mol Neurodegener. 2023. PMID: 37950311 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

