Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway
- PMID: 23843620
- PMCID: PMC3772387
- DOI: 10.1242/jcs.121988
Rac1 is deactivated at integrin activation sites through an IQGAP1-filamin-A-RacGAP1 pathway
Abstract
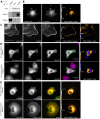
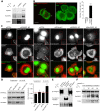
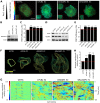
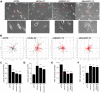
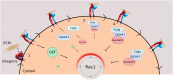
Cell migration makes a fundamental contribution to both normal physiology and disease pathogenesis. Integrin engagement with extracellular ligands spatially controls, via the cyclical activation and deactivation of the small GTPase Rac1, the dynamic membrane protrusion and cytoskeletal reorganization events that are required for directional migration. Although the pathways that control integrin-mediated Rac1 activation are reasonably well defined, the mechanisms that are responsible for switching off activity are poorly understood. Here, proteomic analysis of activated integrin-associated complexes suggests filamin-A and IQ-motif-containing GTPase-activating protein 1 (IQGAP1) as candidates that link β1 integrin to Rac1. siRNA-mediated knockdown of either filamin-A or IQGAP1 induced high, dysregulated Rac1 activity during cell spreading on fibronectin. Using immunoprecipitation and immunocytochemistry, filamin-A and IQGAP1 were shown to be part of a complex that is recruited to active β1 integrin. Mass spectrometric analysis of individual filamin-A, IQGAP1 and Rac1 pull-downs and biochemical analysis, identified RacGAP1 as a novel IQGAP1 binding partner. Further immunoprecipitation and immunocytochemistry analyses demonstrated that RacGAP1 is recruited to IQGAP1 and active β1 integrin, and that suppression of RacGAP1 expression triggered elevated Rac1 activity during spreading on fibronectin. Consistent with these findings, reduced expression of filamin-A, IQGAP1 or RacGAP1 triggered unconstrained membrane protrusion and disrupted directional cell migration on fibrillar extracellular matrices. These findings suggest a model whereby integrin engagement, followed by filamin-A, IQGAP1 and RacGAP1 recruitment, deactivates Rac1 to constrain its activity spatially and thereby coordinate directional cell migration.
Keywords: FLNa; IQGAP1; Integrin; Migration; Rac1; RacGAP1.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

