Contribution of myocyte enhancer factor 2 family transcription factors to BZLF1 expression in Epstein-Barr virus reactivation from latency
- PMID: 23843637
- PMCID: PMC3754021
- DOI: 10.1128/JVI.01002-13
Contribution of myocyte enhancer factor 2 family transcription factors to BZLF1 expression in Epstein-Barr virus reactivation from latency
Abstract
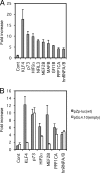
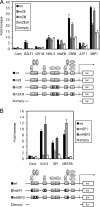
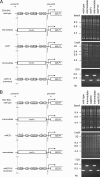
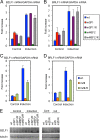
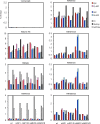
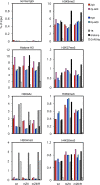
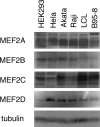
Reactivation of Epstein-Barr virus (EBV) from latency is dependent on expression of the viral transactivator BZLF1 protein, whose promoter (Zp) normally exhibits only low basal activity but is activated in response to chemical or biological inducers. Using a reporter assay system, we screened for factors that can activate Zp and isolated genes, including those encoding MEF2B, KLF4, and some cellular b-Zip family transcription factors. After confirming their importance and functional binding sites in reporter assays, we prepared recombinant EBV-BAC, in which the binding sites were mutated. Interestingly, the MEF2 mutant virus produced very low levels of BRLF1, another transactivator of EBV, in addition to BZLF1 in HEK293 cells. The virus failed to induce a subset of early genes, such as that encoding BALF5, upon lytic induction, and accordingly, could not replicate to produce progeny viruses in HEK293 cells, but this restriction could be completely lifted by exogenous supply of BRLF1, together with BZLF1. In B cells, induction of BZLF1 by chemical inducers was inhibited by point mutations in the ZII or the three SP1/KLF binding sites of EBV-BAC Zp, while leaky BZLF1 expression was less affected. Mutation of MEF2 sites severely impaired both spontaneous and induced expression of not only BZLF1, but also BRLF1 in comparison to wild-type or revertant virus cases. We also observed that MEF2 mutant EBV featured relatively high repressive histone methylation, such as H3K27me3, but CpG DNA methylation levels were comparable around Zp and the BRLF1 promoter (Rp). These findings shed light on BZLF1 expression and EBV reactivation from latency.
Figures





Similar articles
-
Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters.J Virol. 2015 Feb;89(3):1731-43. doi: 10.1128/JVI.02781-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410866 Free PMC article.
-
Viral genome methylation differentially affects the ability of BZLF1 versus BRLF1 to activate Epstein-Barr virus lytic gene expression and viral replication.J Virol. 2013 Jan;87(2):935-50. doi: 10.1128/JVI.01790-12. Epub 2012 Nov 7. J Virol. 2013. PMID: 23135711 Free PMC article.
-
Methylation profiling of Epstein-Barr virus immediate-early gene promoters, BZLF1 and BRLF1 in tumors of epithelial, NK- and B-cell origins.BMC Cancer. 2012 Mar 29;12:125. doi: 10.1186/1471-2407-12-125. BMC Cancer. 2012. PMID: 22458933 Free PMC article.
-
Regulation of Epstein-Barr virus reactivation from latency.Microbiol Immunol. 2014 Jun;58(6):307-17. doi: 10.1111/1348-0421.12155. Microbiol Immunol. 2014. PMID: 24786491 Review.
-
Switching of EBV cycles between latent and lytic states.Rev Med Virol. 2014 May;24(3):142-53. doi: 10.1002/rmv.1780. Epub 2013 Dec 11. Rev Med Virol. 2014. PMID: 24339346 Review.
Cited by
-
The Epstein-Barr Virus BDLF4 Gene Is Required for Efficient Expression of Viral Late Lytic Genes.J Virol. 2015 Oct;89(19):10120-4. doi: 10.1128/JVI.01604-15. Epub 2015 Jul 22. J Virol. 2015. PMID: 26202235 Free PMC article.
-
Cellular differentiation regulator BLIMP1 induces Epstein-Barr virus lytic reactivation in epithelial and B cells by activating transcription from both the R and Z promoters.J Virol. 2015 Feb;89(3):1731-43. doi: 10.1128/JVI.02781-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410866 Free PMC article.
-
Identification of ARKL1 as a Negative Regulator of Epstein-Barr Virus Reactivation.J Virol. 2019 Sep 30;93(20):e00989-19. doi: 10.1128/JVI.00989-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31341047 Free PMC article.
-
Estrogen induces the expression of EBV lytic protein ZEBRA, a marker of poor prognosis in nasopharyngeal carcinoma.Cancer Sci. 2022 Aug;113(8):2862-2877. doi: 10.1111/cas.15440. Epub 2022 Jun 27. Cancer Sci. 2022. PMID: 35633182 Free PMC article.
-
Epstein-Barr Virus Infection of Oral Squamous Cells.Microorganisms. 2020 Mar 16;8(3):419. doi: 10.3390/microorganisms8030419. Microorganisms. 2020. PMID: 32188127 Free PMC article.
References
-
- Tsurumi T, Fujita M, Kudoh A. 2005. Latent and lytic Epstein-Barr virus replication strategies. Rev. Med. Virol. 15:3–15 - PubMed
-
- Amon W, Farrell PJ. 2005. Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15:149–156 - PubMed
-
- Speck SH, Chatila T, Flemington E. 1997. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399–405 - PubMed
-
- Miller G, El-Guindy A, Countryman J, Ye J, Gradoville L. 2007. Lytic cycle switches of oncogenic human gammaherpesviruses. Adv. Cancer Res. 97:81–109 - PubMed
-
- Sinclair AJ. 2003. bZIP proteins of human gammaherpesviruses. J. Gen. Virol. 84:1941–1949 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

