Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii
- PMID: 23843955
- PMCID: PMC3699609
- DOI: 10.1371/journal.pone.0066406
Real-time fluorescence loop mediated isothermal amplification for the detection of Acinetobacter baumannii
Abstract
Background: Detection of Acinetobacter baumannii has been relying primarily on bacterial culture that often fails to return useful results in time. Although DNA-based assays are more sensitive than bacterial culture in detecting the pathogen, the molecular results are often inconsistent and challenged by doubts on false positives, such as those due to system- and environment-derived contaminations. In addition, these molecular tools require expensive laboratory instruments. Therefore, establishing molecular tools for field use require simpler molecular platforms. The loop-mediated isothermal amplification method is relatively simple and can be improved for better use in a routine clinical bacteriology laboratory. A simple and portable device capable of performing both the amplification and detection (by fluorescence) of LAMP in the same platform has been developed in recent years. This method is referred to as real-time loop-mediated isothermal amplification. In this study, we attempted to utilize this method for rapid detection of A. baumannii.
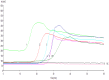
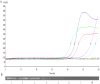
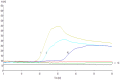
Methodology and significant findings: Species-specific primers were designed to test the utility of this method. Clinical samples of A. baumannii were used to determine the sensitivity and specificity of this system compared to bacterial culture and a polymerase chain reaction method. All positive samples isolated from sputum were confirmed to be the species of Acinetobacter by 16S rRNA gene sequencing. The RealAmp method was found to be simpler and allowed real-time detection of DNA amplification, and could distinguish A. baumannii from Acinetobacter calcoaceticus and Acinetobacter genomic species 3. DNA was extracted by simple boiling method. Compared to bacterial culture, the sensitivity and specificity of RealAmp in detecting A. baumannii was 98.9% and 75.0%, respectively.
Conclusion: The RealAmp assay only requires a single unit, and the assay positivity can be verified by visual inspection. Therefore, this assay has great potential of field use as a molecular tool for detection of A. baumannii.
Conflict of interest statement
Figures
Similar articles
-
Application of LAMP assay for detection of carbapenem-resistant Acinetobacter calcoaceticus-Acinetobacter baumannii complex in ICU admitted sepsis patients: A rapid and cost-effective diagnostic tool.Diagn Microbiol Infect Dis. 2024 Sep;110(1):116398. doi: 10.1016/j.diagmicrobio.2024.116398. Epub 2024 Jun 12. Diagn Microbiol Infect Dis. 2024. PMID: 38908041
-
Oligonucleotide array-based identification of species in the Acinetobacter calcoaceticus-A. baumannii complex in isolates from blood cultures and antimicrobial susceptibility testing of the isolates.J Clin Microbiol. 2008 Jun;46(6):2052-9. doi: 10.1128/JCM.00014-08. Epub 2008 Apr 2. J Clin Microbiol. 2008. PMID: 18385442 Free PMC article.
-
Rapid and sensitive detection of Acinetobacter baumannii using loop-mediated isothermal amplification.J Microbiol Methods. 2013 Feb 15;92(2):197-200. doi: 10.1016/j.mimet.2012.11.020. Epub 2012 Dec 5. J Microbiol Methods. 2013. PMID: 23220188
-
Identification of Acinetobacter isolates in the A. calcoaceticus-A. baumannii complex by restriction analysis of the 16S-23S rRNA intergenic-spacer sequences.J Clin Microbiol. 1995 May;33(5):1108-13. doi: 10.1128/jcm.33.5.1108-1113.1995. J Clin Microbiol. 1995. PMID: 7542263 Free PMC article.
-
Acinetobacter baumannii-calcoaceticus Complex-associated Orbital Cellulitis: A Case Report and Literature Review.Ocul Immunol Inflamm. 2023 Sep;31(7):1537-1540. doi: 10.1080/09273948.2022.2103715. Epub 2022 Sep 8. Ocul Immunol Inflamm. 2023. PMID: 36074653 Review.
Cited by
-
Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms.J Appl Microbiol. 2018 Mar;124(3):626-643. doi: 10.1111/jam.13647. Epub 2018 Feb 12. J Appl Microbiol. 2018. PMID: 29165905 Free PMC article. Review.
-
Rapid detection of Streptococcus pneumoniae by real-time fluorescence loop-mediated isothermal amplification.J Thorac Dis. 2014 Sep;6(9):1193-9. doi: 10.3978/j.issn.2072-1439.2014.07.29. J Thorac Dis. 2014. PMID: 25276360 Free PMC article.
-
Rapid detection of Acinetobacter baumannii and molecular epidemiology of carbapenem-resistant A. baumannii in two comprehensive hospitals of Beijing, China.Front Microbiol. 2015 Sep 23;6:997. doi: 10.3389/fmicb.2015.00997. eCollection 2015. Front Microbiol. 2015. PMID: 26441924 Free PMC article.
-
Development of Loop-Mediated Isothermal Amplification Assay for Detection of Clinically Significant Members of Acinetobacter calcoaceticus-baumannii Complex and Associated Carbapenem Resistance.Front Mol Biosci. 2021 Jun 23;8:659256. doi: 10.3389/fmolb.2021.659256. eCollection 2021. Front Mol Biosci. 2021. PMID: 34250011 Free PMC article.
-
Point-of-Care Diagnostic Testing for Emerging and Existing Poultry Viral Respiratory Pathogens Using Loop-Mediated Isothermal Amplification.Pathogens. 2025 Jul 2;14(7):657. doi: 10.3390/pathogens14070657. Pathogens. 2025. PMID: 40732704 Free PMC article. Review.
References
-
- Visca P, Seifert H, Towner KJ (2011) Acinetobacter infection – an emerging threat to human health. 63: 1048–1054. - PubMed
-
- Chen TL, Siu LK, Wu RC, Shaio MF, Huang LY, et al. (2007) Comparison of one-tube multiplex PCR, automated ribotyping and intergenic spacer (ITS) sequencing for rapid identification of Acinetobacter baumannii Clin Microbiol Infect. 13: 801–806. - PubMed
-
- Higgins PG, Wisplinghoff H, Krut O, Seifert H (2007) A PCR-based method to differentiate between Acinetobacter baumannii and Acinetobacter genomic species 13TU. Clin Microbiol Infect 13: 1199–1201. - PubMed
-
- García-Arata MI, Gerner-Smidt P, Baquero F, Ibrahim A (1997) PCR-amplified 16S and 23S rDNA restriction analysis for the identification of Acinetobacter strains at the DNA group level. Res Microbiol 148: 777–784. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

