Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays
- PMID: 23843967
- PMCID: PMC3700933
- DOI: 10.1371/journal.pone.0066843
Unravelling the complexity of human olfactory receptor repertoire by copy number analysis across population using high resolution arrays
Abstract
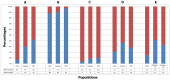
Olfactory receptors (OR), responsible for detection of odor molecules, belong to the largest family of genes and are highly polymorphic in nature having distinct polymorphisms associated with specific regions around the globe. Since there are no reports on the presence of copy number variations in OR repertoire of Indian population, the present investigation in 43 Indians along with 270 HapMap and 31 Tibetan samples was undertaken to study genome variability and evolution. Analysis was performed using Affymetrix Genome-Wide Human SNP Array 6.0 chip, Affymterix CytoScan(®) High-Density array, HD-CNV, and MAFFT program. We observed a total of 1527 OR genes in 503 CNV events from 81.3% of the study group, which includes 67.6% duplications and 32.4% deletions encompassing more of genes than pseudogenes. We report human genotypic variation in functional OR repertoire size across populations and it was found that the combinatorial effect of both "orthologous obtained from closely related species" and "paralogous derived sequences" provide the complexity to the continuously occurring OR CNVs.
Conflict of interest statement
Figures




Similar articles
-
Global patterns of large copy number variations in the human genome reveal complexity in chromosome organization.Genet Res (Camb). 2015 Sep 22;97:e18. doi: 10.1017/S0016672315000191. Genet Res (Camb). 2015. PMID: 26390810 Free PMC article.
-
High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution.PLoS Genet. 2008 Nov;4(11):e1000249. doi: 10.1371/journal.pgen.1000249. Epub 2008 Nov 7. PLoS Genet. 2008. PMID: 18989455 Free PMC article.
-
Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity.PLoS Comput Biol. 2010 Nov 11;6(11):e1000988. doi: 10.1371/journal.pcbi.1000988. PLoS Comput Biol. 2010. PMID: 21085617 Free PMC article.
-
Human olfaction: from genomic variation to phenotypic diversity.Trends Genet. 2009 Apr;25(4):178-84. doi: 10.1016/j.tig.2009.02.002. Epub 2009 Mar 18. Trends Genet. 2009. PMID: 19303166 Review.
-
Odor discrimination by G protein-coupled olfactory receptors.Microsc Res Tech. 2002 Aug 1;58(3):135-41. doi: 10.1002/jemt.10131. Microsc Res Tech. 2002. PMID: 12203691 Review.
Cited by
-
Genome-wide copy number scan identifies IRF6 involvement in Van der Woude syndrome in an Indian family.Genet Res (Camb). 2014 Oct 10;96:e12. doi: 10.1017/S0016672314000159. Genet Res (Camb). 2014. PMID: 25579819 Free PMC article.
-
Advancements in copy number variation screening in herbivorous livestock genomes and their association with phenotypic traits.Front Vet Sci. 2024 Jan 11;10:1334434. doi: 10.3389/fvets.2023.1334434. eCollection 2023. Front Vet Sci. 2024. PMID: 38274664 Free PMC article. Review.
-
Copy number variation burden on asthma subgenome in normal cohorts identifies susceptibility markers.Allergy Asthma Immunol Res. 2015 May;7(3):265-75. doi: 10.4168/aair.2015.7.3.265. Epub 2014 Dec 18. Allergy Asthma Immunol Res. 2015. PMID: 25749760 Free PMC article.
-
Global patterns of large copy number variations in the human genome reveal complexity in chromosome organization.Genet Res (Camb). 2015 Sep 22;97:e18. doi: 10.1017/S0016672315000191. Genet Res (Camb). 2015. PMID: 26390810 Free PMC article.
-
Global Survey of Variation in a Human Olfactory Receptor Gene Reveals Signatures of Non-Neutral Evolution.Chem Senses. 2015 Sep;40(7):481-8. doi: 10.1093/chemse/bjv030. Epub 2015 Jun 13. Chem Senses. 2015. PMID: 26072518 Free PMC article.
References
-
- Hasin-Brumshtein Y, Lancet D, Olender T (2009) Human olfaction: from genomic variation to phenotypic diversity. Trends Genet 25(4): 178–184. - PubMed
-
- Fuchs T, Glusman G, Horn-Saban S, Lancet D, Pilpel Y (2001) The human olfactory subgenome: from sequence to structure and evolution. Hum Genet 108(1): 1–13. - PubMed
-
- Freeman JL, Perry GH, Feuk L, Redon R, McCarroll SA, et al. (2006) Copy number variation: New insights in genome diversity. Genome Res 16: 949–961. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

