Using high-throughput sequencing to leverage surveillance of genetic diversity and oseltamivir resistance: a pilot study during the 2009 influenza A(H1N1) pandemic
- PMID: 23843978
- PMCID: PMC3699567
- DOI: 10.1371/journal.pone.0067010
Using high-throughput sequencing to leverage surveillance of genetic diversity and oseltamivir resistance: a pilot study during the 2009 influenza A(H1N1) pandemic
Abstract
Background: Influenza viruses display a high mutation rate and complex evolutionary patterns. Next-generation sequencing (NGS) has been widely used for qualitative and semi-quantitative assessment of genetic diversity in complex biological samples. The "deep sequencing" approach, enabled by the enormous throughput of current NGS platforms, allows the identification of rare genetic viral variants in targeted genetic regions, but is usually limited to a small number of samples.
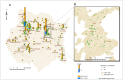
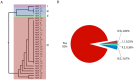
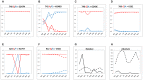
Methodology and principal findings: We designed a proof-of-principle study to test whether redistributing sequencing throughput from a high depth-small sample number towards a low depth-large sample number approach is feasible and contributes to influenza epidemiological surveillance. Using 454-Roche sequencing, we sequenced at a rather low depth, a 307 bp amplicon of the neuraminidase gene of the Influenza A(H1N1) pandemic (A(H1N1)pdm) virus from cDNA amplicons pooled in 48 barcoded libraries obtained from nasal swab samples of infected patients (n = 299) taken from May to November, 2009 pandemic period in Mexico. This approach revealed that during the transition from the first (May-July) to second wave (September-November) of the pandemic, the initial genetic variants were replaced by the N248D mutation in the NA gene, and enabled the establishment of temporal and geographic associations with genetic diversity and the identification of mutations associated with oseltamivir resistance.
Conclusions: NGS sequencing of a short amplicon from the NA gene at low sequencing depth allowed genetic screening of a large number of samples, providing insights to viral genetic diversity dynamics and the identification of genetic variants associated with oseltamivir resistance. Further research is needed to explain the observed replacement of the genetic variants seen during the second wave. As sequencing throughput rises and library multiplexing and automation improves, we foresee that the approach presented here can be scaled up for global genetic surveillance of influenza and other infectious diseases.
Conflict of interest statement
Figures



Similar articles
-
Genetic sequencing of influenza A (H1N1) pdm09 isolates from South India, collected between 2011 and 2015 to detect mutations affecting virulence and resistance to oseltamivir.Indian J Med Microbiol. 2020 Jul-Dec;38(3 & 4):324-337. doi: 10.4103/ijmm.IJMM_20_83. Indian J Med Microbiol. 2020. PMID: 33154243
-
Mutation analysis of 2009 pandemic influenza A(H1N1) viruses collected in Japan during the peak phase of the pandemic.PLoS One. 2011 Apr 29;6(4):e18956. doi: 10.1371/journal.pone.0018956. PLoS One. 2011. PMID: 21572517 Free PMC article.
-
Genetic diversity among pandemic 2009 influenza viruses isolated from a transmission chain.Virol J. 2013 Apr 12;10:116. doi: 10.1186/1743-422X-10-116. Virol J. 2013. PMID: 23587185 Free PMC article.
-
Emerging oseltamivir resistance in seasonal and pandemic influenza A/H1N1.J Clin Virol. 2011 Oct;52(2):70-8. doi: 10.1016/j.jcv.2011.05.019. Epub 2011 Jun 17. J Clin Virol. 2011. PMID: 21684202 Review.
-
Emergence of oseltamivir resistance: control and management of influenza before, during and after the pandemic.Infect Disord Drug Targets. 2013 Feb;13(1):34-45. doi: 10.2174/18715265112129990006. Infect Disord Drug Targets. 2013. PMID: 23675925 Review.
Cited by
-
Clinical Implications of Antiviral Resistance in Influenza.Viruses. 2015 Sep 14;7(9):4929-44. doi: 10.3390/v7092850. Viruses. 2015. PMID: 26389935 Free PMC article. Review.
-
Comparison of three next-generation sequencing platforms for metagenomic sequencing and identification of pathogens in blood.BMC Genomics. 2014 Feb 4;15:96. doi: 10.1186/1471-2164-15-96. BMC Genomics. 2014. PMID: 24495417 Free PMC article.
-
Deep sequencing: becoming a critical tool in clinical virology.J Clin Virol. 2014 Sep;61(1):9-19. doi: 10.1016/j.jcv.2014.06.013. Epub 2014 Jun 24. J Clin Virol. 2014. PMID: 24998424 Free PMC article. Review.
-
Seasonal Genetic Drift of Human Influenza A Virus Quasispecies Revealed by Deep Sequencing.Front Microbiol. 2018 Oct 31;9:2596. doi: 10.3389/fmicb.2018.02596. eCollection 2018. Front Microbiol. 2018. PMID: 30429836 Free PMC article.
-
Antiviral resistance markers in influenza virus sequences in Mexico, 2000-2017.Infect Drug Resist. 2018 Oct 11;11:1751-1756. doi: 10.2147/IDR.S153154. eCollection 2018. Infect Drug Resist. 2018. PMID: 30349332 Free PMC article.
References
-
- Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, et al. (2012) Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 12: 687–695. - PubMed
-
- Pizzorno A, Abed Y, Boivin G (2011) Influenza drug resistance. Semin Respir Crit Care Med 32: 409–422. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

