High throughput gene expression analysis identifies reliable expression markers of human corneal endothelial cells
- PMID: 23844023
- PMCID: PMC3699644
- DOI: 10.1371/journal.pone.0067546
High throughput gene expression analysis identifies reliable expression markers of human corneal endothelial cells
Abstract
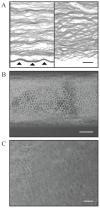
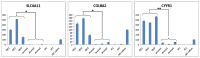
Considerable interest has been generated for the development of suitable corneal endothelial graft alternatives through cell-tissue engineering, which can potentially alleviate the shortage of corneal transplant material. The advent of less invasive suture-less key-hole surgery options such as Descemet's Stripping Endothelial Keratoplasty (DSEK) and Descemet's Membrane Endothelial Keratoplasty (DMEK), which involve transplantation of solely the endothelial layer instead of full thickness cornea, provide further impetus for the development of alternative endothelial grafts for clinical applications. A major challenge for this endeavor is the lack of specific markers for this cell type. To identify genes that reliably mark corneal endothelial cells (CECs) in vivo and in vitro, we performed RNA-sequencing on freshly isolated human CECs (from both young and old donors), CEC cultures, and corneal stroma. Gene expression of these corneal cell types was also compared to that of other human tissue types. Based on high throughput comparative gene expression analysis, we identified a panel of markers that are: i) highly expressed in CECs from both young donors and old donors; ii) expressed in CECs in vivo and in vitro; and iii) not expressed in corneal stroma keratocytes and the activated corneal stroma fibroblasts. These were SLC4A11, COL8A2 and CYYR1. The use of this panel of genes in combination reliably ascertains the identity of the CEC cell type.
Conflict of interest statement
Figures
References
-
- Peh GS, Beuerman RW, Colman A, Tan DT, Mehta JS (2011) Human corneal endothelial cell expansion for corneal endothelium transplantation: an overview. Transplantation 91: 811–819. - PubMed
-
- Bourne WM, Nelson LR, Hodge DO (1997) Central corneal endothelial cell changes over a ten-year period. Investigative ophthalmology & visual science 38: 779–782. - PubMed
-
- Joyce NC (2003) Proliferative capacity of the corneal endothelium. Progress in retinal and eye research 22: 359–389. - PubMed
-
- Kaufman HE, Katz JI (1977) Pathology of the corneal endothelium. Investigative ophthalmology & visual science 16: 265–268. - PubMed
-
- Edelhauser HF (2000) The resiliency of the corneal endothelium to refractive and intraocular surgery. Cornea 19: 263–273. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

