The mode of inhibitor binding to peptidyl-tRNA hydrolase: binding studies and structure determination of unbound and bound peptidyl-tRNA hydrolase from Acinetobacter baumannii
- PMID: 23844024
- PMCID: PMC3701073
- DOI: 10.1371/journal.pone.0067547
The mode of inhibitor binding to peptidyl-tRNA hydrolase: binding studies and structure determination of unbound and bound peptidyl-tRNA hydrolase from Acinetobacter baumannii
Abstract
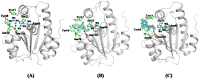
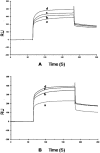
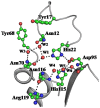
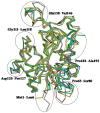
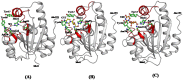
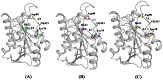
The incidences of infections caused by an aerobic Gram-negative bacterium, Acinetobacter baumannii are very common in hospital environments. It usually causes soft tissue infections including urinary tract infections and pneumonia. It is difficult to treat due to acquired resistance to available antibiotics is well known. In order to design specific inhibitors against one of the important enzymes, peptidyl-tRNA hydrolase from Acinetobacter baumannii, we have determined its three-dimensional structure. Peptidyl-tRNA hydrolase (AbPth) is involved in recycling of peptidyl-tRNAs which are produced in the cell as a result of premature termination of translation process. We have also determined the structures of two complexes of AbPth with cytidine and uridine. AbPth was cloned, expressed and crystallized in unbound and in two bound states with cytidine and uridine. The binding studies carried out using fluorescence spectroscopic and surface plasmon resonance techniques revealed that both cytidine and uridine bound to AbPth at nanomolar concentrations. The structure determinations of the complexes revealed that both ligands were located in the active site cleft of AbPth. The introduction of ligands to AbPth caused a significant widening of the entrance gate to the active site region and in the process of binding, it expelled several water molecules from the active site. As a result of interactions with protein atoms, the ligands caused conformational changes in several residues to attain the induced tight fittings. Such a binding capability of this protein makes it a versatile molecule for hydrolysis of peptidyl-tRNAs having variable peptide sequences. These are the first studies that revealed the mode of inhibitor binding in Peptidyl-tRNA hydrolases which will facilitate the structure based ligand design.
Conflict of interest statement
Figures




Similar articles
-
Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.Biochem J. 2018 Feb 9;475(3):547-560. doi: 10.1042/BCJ20170666. Biochem J. 2018. PMID: 29301982
-
Structural and binding studies of peptidyl-tRNA hydrolase from Pseudomonas aeruginosa provide a platform for the structure-based inhibitor design against peptidyl-tRNA hydrolase.Biochem J. 2014 Nov 1;463(3):329-37. doi: 10.1042/BJ20140631. Biochem J. 2014. PMID: 25101795
-
Crystal structure at 1.2 A resolution and active site mapping of Escherichia coli peptidyl-tRNA hydrolase.EMBO J. 1997 Aug 1;16(15):4760-9. doi: 10.1093/emboj/16.15.4760. EMBO J. 1997. PMID: 9303320 Free PMC article.
-
Structural and functional insights into peptidyl-tRNA hydrolase.Biochim Biophys Acta. 2014 Jul;1844(7):1279-88. doi: 10.1016/j.bbapap.2014.04.012. Epub 2014 Apr 21. Biochim Biophys Acta. 2014. PMID: 24768774 Review.
-
Unveiling the Druggable Landscape of Bacterial Peptidyl tRNA Hydrolase: Insights into Structure, Function, and Therapeutic Potential.Biomolecules. 2024 Jun 7;14(6):668. doi: 10.3390/biom14060668. Biomolecules. 2024. PMID: 38927071 Free PMC article. Review.
Cited by
-
Miscoding-induced stalling of substrate translocation on the bacterial ribosome.Proc Natl Acad Sci U S A. 2017 Oct 10;114(41):E8603-E8610. doi: 10.1073/pnas.1707539114. Epub 2017 Sep 25. Proc Natl Acad Sci U S A. 2017. PMID: 28973849 Free PMC article.
-
Crystal structure of peptidyl-tRNA hydrolase from a Gram-positive bacterium, Streptococcus pyogenes at 2.19 Å resolution shows the closed structure of the substrate-binding cleft.FEBS Open Bio. 2014 Oct 22;4:915-22. doi: 10.1016/j.fob.2014.10.010. eCollection 2014. FEBS Open Bio. 2014. PMID: 25389518 Free PMC article.
-
Small Molecule Docking Supports Broad and Narrow Spectrum Potential for the Inhibition of the Novel Antibiotic Target Bacterial Pth1.Antibiotics (Basel). 2016 May 10;5(2):16. doi: 10.3390/antibiotics5020016. Antibiotics (Basel). 2016. PMID: 27171117 Free PMC article.
-
Expedited isolation of natural product peptidyl-tRNA hydrolase inhibitors from a Pth1 affinity column.AIMS Mol Sci. 2017;4(2):175-184. doi: 10.3934/molsci.2017.2.175. Epub 2017 May 12. AIMS Mol Sci. 2017. PMID: 30740515 Free PMC article.
-
Unraveling the stereochemical and dynamic aspects of the catalytic site of bacterial peptidyl-tRNA hydrolase.RNA. 2017 Feb;23(2):202-216. doi: 10.1261/rna.057620.116. Epub 2016 Nov 10. RNA. 2017. PMID: 28096445 Free PMC article.
References
-
- Das G, Varshney U (2006) Peptidyl tRNA hydrolase and its critical role in protein biosynthesis. Microbiology 152: 2191–2195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

