Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae
- PMID: 23844123
- PMCID: PMC3699464
- DOI: 10.1371/journal.pone.0067902
Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae
Abstract
Fluorescent protein fusions are a powerful tool to monitor the localization and trafficking of proteins. Such studies are particularly easy to carry out in the budding yeast Saccharomyces cerevisiae due to the ease with which tags can be introduced into the genome by homologous recombination. However, the available yeast tagging plasmids have not kept pace with the development of new and improved fluorescent proteins. Here, we have constructed yeast optimized versions of 19 different fluorescent proteins and tested them for use as fusion tags in yeast. These include two blue, seven green, and seven red fluorescent proteins, which we have assessed for brightness, photostability and perturbation of tagged proteins. We find that EGFP remains the best performing green fluorescent protein, that TagRFP-T and mRuby2 outperform mCherry as red fluorescent proteins, and that mTagBFP2 can be used as a blue fluorescent protein tag. Together, the new tagging vectors we have constructed provide improved blue and red fluorescent proteins for yeast tagging and three color imaging.
Conflict of interest statement
Figures

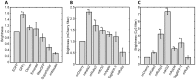
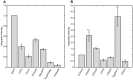
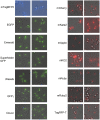

References
-
- Petracek ME, Longtine MS (2002) PCR-based engineering of yeast genome. Meth Enzymol 350: 445–469. - PubMed
-
- Slaughter BD, Schwartz JW, Li R (2007) Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proc Natl Acad Sci USA 104: 20320–20325 doi:10.1073/pnas.0710336105 - DOI - PMC - PubMed
-
- Hailey DW, Davis TN, Muller EGD (2002) Fluorescence resonance energy transfer using color variants of green fluorescent protein. Meth Enzymol 351: 34–49. - PubMed
-
- Reid RJD, Lisby M, Rothstein R (2002) Cloning-free genome alterations in Saccharomyces cerevisiae using adaptamer-mediated PCR. Meth Enzymol 350: 258–277. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

