Associations of ATR and CHEK1 single nucleotide polymorphisms with breast cancer
- PMID: 23844225
- PMCID: PMC3700940
- DOI: 10.1371/journal.pone.0068578
Associations of ATR and CHEK1 single nucleotide polymorphisms with breast cancer
Abstract
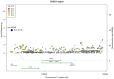
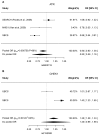
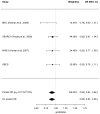
DNA damage and replication checkpoints mediated by the ATR-CHEK1 pathway are key to the maintenance of genome stability, and both ATR and CHEK1 have been proposed as potential breast cancer susceptibility genes. Many novel variants recently identified by the large resequencing projects have not yet been thoroughly tested in genome-wide association studies for breast cancer susceptibility. We therefore used a tagging SNP (tagSNP) approach based on recent SNP data available from the 1000 genomes projects, to investigate the roles of ATR and CHEK1 in breast cancer risk and survival. ATR and CHEK1 tagSNPs were genotyped in the Sheffield Breast Cancer Study (SBCS; 1011 cases and 1024 controls) using Illumina GoldenGate assays. Untyped SNPs were imputed using IMPUTE2, and associations between genotype and breast cancer risk and survival were evaluated using logistic and Cox proportional hazard regression models respectively on a per allele basis. Significant associations were further examined in a meta-analysis of published data or confirmed in the Utah Breast Cancer Study (UBCS). The most significant associations for breast cancer risk in SBCS came from rs6805118 in ATR (p=7.6 x 10(-5)) and rs2155388 in CHEK1 (p=3.1 x 10(-6)), but neither remained significant after meta-analysis with other studies. However, meta-analysis of published data revealed a weak association between the ATR SNP rs1802904 (minor allele frequency is 12%) and breast cancer risk, with a summary odds ratio (confidence interval) of 0.90 (0.83-0.98) [p=0.0185] for the minor allele. Further replication of this SNP in larger studies is warranted since it is located in the target region of 2 microRNAs. No evidence of any survival effects of ATR or CHEK1 SNPs were identified. We conclude that common alleles of ATR and CHEK1 are not implicated in breast cancer risk or survival, but we cannot exclude effects of rare alleles and of common alleles with very small effect sizes.
Conflict of interest statement
Figures

References
-
- Stratton MR, Rahman N (2008) The emerging landscape of breast cancer susceptibility. Nat Genet 40: 17-22. doi:10.1038/ng.2007.53. PubMed: 18163131. - DOI - PubMed
-
- Houlston RS, Webb E, Broderick P, Pittman AM, Di Bernardo MC et al. (2008) Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet 40: 1426-1435. doi:10.1038/ng.262. PubMed: 19011631. - DOI - PMC - PubMed
-
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL et al. (2008) Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40: 638-645. doi:10.1038/ng.120. PubMed: 18372903. - DOI - PMC - PubMed
-
- Altshuler DL, Durbin RM, Abecasis GR, Bentley DR, Chakravarti A et al. (2010) A map of human genome variation from population-scale sequencing. Nature 467: 1061-1073. doi:10.1038/nature09534. PubMed: 20981092. - DOI - PMC - PubMed
-
- Enders GH (2008) Expanded roles for Chk1 in genome maintenance. J Biol Chem 283: 17749-17752. doi:10.1074/jbc.R800021200. PubMed: 18424430. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

