Glossina fuscipes populations provide insights for human African trypanosomiasis transmission in Uganda
- PMID: 23845311
- PMCID: PMC3772539
- DOI: 10.1016/j.pt.2013.06.005
Glossina fuscipes populations provide insights for human African trypanosomiasis transmission in Uganda
Abstract
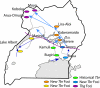
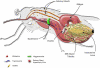
Uganda has both forms of human African trypanosomiasis (HAT): the chronic gambiense disease in the northwest and the acute rhodesiense disease in the south. The recent spread of rhodesiense into central Uganda has raised concerns given the different control strategies the two diseases require. We present knowledge on the population genetics of the major vector species Glossina fuscipes fuscipes in Uganda with a focus on population structure, measures of gene flow between populations, and the occurrence of polyandry. The microbiome composition and diversity is discussed, focusing on their potential role on trypanosome infection outcomes. We discuss the implications of these findings for large-scale tsetse control programs, including suppression or eradication, being undertaken in Uganda, and potential future genetic applications.
Keywords: Glossina fuscipes; Trypanosoma brucei gambiense; Trypanosoma brucei rhodesiense; Uganda; human African trypanosomiasis; population genetics; sleeping sickness; vector control.
Copyright © 2013. Published by Elsevier Ltd.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

