Cancer heterogeneity--a multifaceted view
- PMID: 23846313
- PMCID: PMC3736134
- DOI: 10.1038/embor.2013.92
Cancer heterogeneity--a multifaceted view
Abstract
Cancers of various organs have been categorized into distinct subtypes after increasingly sophisticated taxonomies. Additionally, within a seemingly homogeneous subclass, individual cancers contain diverse tumour cell populations that vary in important cancer-specific traits such as clonogenicity and invasive potential. Differences that exist between and within a given tumour type have hampered significantly both the proper selection of patients that might benefit from therapy, as well as the development of new targeted agents. In this review, we discuss the differences associated with organ-specific cancer subtypes and the factors that contribute to intra-tumour heterogeneity. It is of utmost importance to understand the biological causes that distinguish tumours as well as distinct tumour cell populations within malignancies, as these will ultimately point the way to more rational anti-cancer treatments.
Conflict of interest statement
The authors declare that they have no conflict of interest.
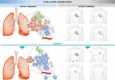
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

