JBioWH: an open-source Java framework for bioinformatics data integration
- PMID: 23846595
- PMCID: PMC3708619
- DOI: 10.1093/database/bat051
JBioWH: an open-source Java framework for bioinformatics data integration
Abstract
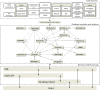
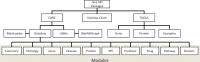
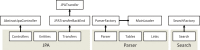
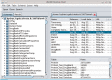
The Java BioWareHouse (JBioWH) project is an open-source platform-independent programming framework that allows a user to build his/her own integrated database from the most popular data sources. JBioWH can be used for intensive querying of multiple data sources and the creation of streamlined task-specific data sets on local PCs. JBioWH is based on a MySQL relational database scheme and includes JAVA API parser functions for retrieving data from 20 public databases (e.g. NCBI, KEGG, etc.). It also includes a client desktop application for (non-programmer) users to query data. In addition, JBioWH can be tailored for use in specific circumstances, including the handling of massive queries for high-throughput analyses or CPU intensive calculations. The framework is provided with complete documentation and application examples and it can be downloaded from the Project Web site at http://code.google.com/p/jbiowh. A MySQL server is available for demonstration purposes at hydrax.icgeb.trieste.it:3307. Database URL: http://code.google.com/p/jbiowh.
Figures

References
-
- Stein LD. Integrating biological databases. Nat. Rev. Genet. 2003;4:337–345. - PubMed
-
- Toomula N, Kumar A, Kumar DS, et al. Biological databases-integration of life science data. J. Comput. Sci. Syst. Biol. 2012;04:87–92.
-
- Anguita A, Martín L, Pérez-Rey D, et al. A review of methods and tools for database integration in biomedicine. Curr. Bioinform. 2010;5:253–269.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

