A compendium of RNA-binding motifs for decoding gene regulation
- PMID: 23846655
- PMCID: PMC3929597
- DOI: 10.1038/nature12311
A compendium of RNA-binding motifs for decoding gene regulation
Abstract
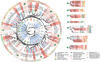
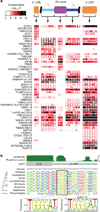
RNA-binding proteins are key regulators of gene expression, yet only a small fraction have been functionally characterized. Here we report a systematic analysis of the RNA motifs recognized by RNA-binding proteins, encompassing 205 distinct genes from 24 diverse eukaryotes. The sequence specificities of RNA-binding proteins display deep evolutionary conservation, and the recognition preferences for a large fraction of metazoan RNA-binding proteins can thus be inferred from their RNA-binding domain sequence. The motifs that we identify in vitro correlate well with in vivo RNA-binding data. Moreover, we can associate them with distinct functional roles in diverse types of post-transcriptional regulation, enabling new insights into the functions of RNA-binding proteins both in normal physiology and in human disease. These data provide an unprecedented overview of RNA-binding proteins and their targets, and constitute an invaluable resource for determining post-transcriptional regulatory mechanisms in eukaryotes.
Figures

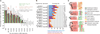

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- DK015602-05/DK/NIDDK NIH HHS/United States
- MOP-93671/CAPMC/ CIHR/Canada
- R01 CA104708/CA/NCI NIH HHS/United States
- MOP-67011/CAPMC/ CIHR/Canada
- MOP-49451/CAPMC/ CIHR/Canada
- P30 CA014520/CA/NCI NIH HHS/United States
- R01 GM051968/GM/NIGMS NIH HHS/United States
- MOP-14409/CAPMC/ CIHR/Canada
- 1R01HG00570/HG/NHGRI NIH HHS/United States
- T32 GM008061/GM/NIGMS NIH HHS/United States
- R01 GM084034/GM/NIGMS NIH HHS/United States
- R01 GM058728/GM/NIGMS NIH HHS/United States
- MOP-125894/CAPMC/ CIHR/Canada
- R01GM084034/GM/NIGMS NIH HHS/United States
- Z01 DK015602/ImNIH/Intramural NIH HHS/United States
- R01 GM114386/GM/NIGMS NIH HHS/United States
- R01 HG005700/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

